-
野油菜黄单胞菌野油菜致病变种(Xanthomonas campestris pv. campestris,Xcc)是十字花科黑腐病的病原菌,通过水孔或气孔进入十字花科植物维管束,并在此定殖、扩散[1]。病原菌生长及侵染受各类蛋白质合成和降解的调控,蛋白酶是调控蛋白降解和激活蛋白活性的重要因素[2]。肽酶在水解肽键降解蛋白质中起到非常重要的作用,根据其催化的反应,肽酶可以分为内肽酶、氨基肽酶和羧基肽酶[3]。脯氨酸亚氨基肽酶(Proline Imino-Peptidase, PIPase)负责从多肽或蛋白质中去除N端脯氨酸残基[3 − 4],也可以去除N端L-丙氨酸或D-丙氨酸,但效率较低[5]。在食品工业中,PIPase用于去除奶酪加工中的肽和含脯氨酸的二肽[6 − 8]。在Xcc中,PipA具有PIPase活性,是其致病力和运动能力的重要调节因子[9]。PipA在许多植物病原菌中高度保守,暗示其可能是这些病原菌致病力的重要调控因子,但Xcc中是否还有其他蛋白也具有PIPase活性并不清楚。
本研究通过体外酶活分析鉴定了Xcc中另一个PIPase蛋白XC3394(PipB),比较了PipA和PipB对Xcc致病力的调控能力,发现PipA和PipB均正调控Xcc的致病力,负调控Xcc的运动性;PipA调控病原菌胞外蛋白酶活性,但PipB没有此调控功能。此外,PipB不能互补PipA的功能,2个PIPase在Xcc中的调控通路有差异,并不是功能冗余的。该研究为解析PIPase在Xcc中的作用机制奠定了基础,为更加全面认识病原菌与宿主互作提供了新思路。
-
本实验所使用的菌株和质粒如表1所示。
表 1 菌株和质粒来源
Table 1. Strains and plasmids used in this study
名称Name 相关特性Correlation characteristic 来源Source Strains Xcc 8004 A wild-type Xcc strain, a laboratory strain with spontaneous rifampicin-resistance Lab collections DH5α Genetically engineered recipient bacteria. Biomed BL21(DE3) Bacteria for protein expression Biomed ∆pipA The pipA deletion mutant of WT This study ∆pipB The pipB deletion mutant of WT This study ∆pipA/∆pipB The pipA and pipB deletion mutant of WT This study pipA/∆pipA The pipA complementation strain of ∆pipA This study pipB/∆pipB The pipB complementation strain of ∆pipB This study pipA/∆pipB The pipA complementation strain of ∆pipB This study pipB/∆pipA The pipB complementation strain of ∆pipA This study pipA/∆pipA/∆pipB The pipA complementation strain of ∆pipA/∆pipB This study pipB/∆pipA/∆pipB The pipB complementation strain of ∆pipA/∆pipB This study Plasmids pK18mobSacB Xanthomonas suicide vector, KanR [10] pHMI Broad host vector, SpR [11] pQE80L Protein expression vector, AmpR Qiagen pK18-pipA pK18mobSacB based plasmid for pipA deletion This study pK18-pipB pK18mobSacB based plasmid for pipB deletion This study pHMI-pipA pHMI based plasmid for pipA expression This study pHMI-pipB pHMI based plasmid for pipB expression This study pQE80L-pipA pQE80L based plasmid for pipA expression in E. coli This study pQE80L-pipB pQE80L based plasmid for pipB expression in E. coli This study -
实验所用引物详见表2。
表 2 本研究所用引物
Table 2. Primers used in this study
引物Primer 序列Sequence(5′-3′) 用途Usages pipAddFF TCTGAAGCTTGCTGCTGGAGTTGTAGGAAG pipA deletion pipAddFR TCTGGGATCCGATGCGGTACCAGGTGCGAT pipAddRF ACGTGGATCCAACTTCCTCGACGACCACAG pipAddRR ACTCGAATTCAACGTAGTGCGCAGCACTAC pipBddFF TCTGAAGCTTGAGCGGTTCGATCTGATCGC pipB deletion pipBddFR TCTGGGATCCCATGCGTCGGTCCTGCGGGA pipBddRF ACGTGGATCCGACAGCTTCGCCTGAGAAGG pipBddRR ACTCGAATTCATCGGCTGCTGCAGGCTTTG pipAhmF ACCATGATTACGCCAAGCTTGATGCGCACGCTCTATCCCGA ΔpipA complementation pipAhmR GTAGAATTCTAGAGGGTACCAGACGGCACGATTGGCCAGCG pipBhmF ACCATGATTACGCCAAGCTTGATGCGCACGCTCTATCCCGA ΔpipB complementation pipBhmR GTAGAATTCTAGAGGGTACCAGGCGAAGCTGTCGGTCGCAC pipAqeF ACGTGGATCCATGCAGTGCACCGAGGGTTTC PipA expression pipAqeR ACGCAAGCTTAGACGGCACGATTGGCCAGCG pipBqeF ACGTGGATCCATGCGCACGCTCTATCCCGAG PipB expression pipBqeR ACGCAAGCTTAGGCGAAGCTGTCGGTCGCAC 注:下划线表示酶切位点。
Note: The enzyme cutting sites are underlined. -
以Xcc
8004 基因组为模板,以pipAqeF/pipAqeR、pipBqeF/pipBqeR为引物,扩增pipA和pipB基因片段,BamHI/HindⅢ酶切后连接到相同酶切的pQE80L载体上,测序验证正确后获得His-PipA、His-PipB融合表达载体pQE80L-pipA、pQE80L-pipB,然后将融合表达载体转化BL21(DE3)细胞[12]。菌株37 ℃过夜培养后,以1∶100体积比转接到500 mL新鲜溶菌肉汤(Luria Bertani, LB)液体培养基继续培养至OD600值达0.6~0.8后,加入250 µL 1 mol·L−1异丙基-β-D-硫代半乳糖苷(Isopropyl-beta-D-thiogalactopyranoside, IPTG),并于25 ℃、90 r·min−1条件下诱导培养5~6 h。离心收集菌体后进行超声破碎(超声功率比40%、超声5 s、间隙5 s、总时间10 min),取上清液进行SDS-PAGE电泳观察蛋白诱导表达情况。利用His标签蛋白纯化蛋白原理,采用His-tag纯化磁珠(BeaverBeads®IDA-Nikel Kit)与上清进行共孵育,经过低浓度咪唑(50 mmol·L−1)洗杂再用高浓度咪唑(500 mmol·L−1)洗脱蛋白,最后用蛋白浓缩柱(Millipore)将洗脱蛋白进行浓缩并置换至50 mmol·L−1 Tris-HCl(pH8.0)缓冲液中,按照SDS-PAGE电泳结果标定一致蛋白量,取相同蛋白量的2 µL进行酶活力验证实验。 -
以对硝基苯胺(p-nitroanilide, pNA)作为底物测定PipA和PipB的肽酶活性。以MBP蛋白作为空白对照,以无蛋白体系为阴性对照。使用分光光度计在405 nm处测量吸光值。反应体系如下:1 mmol·L−1 Tris-HCl (pH8.0)、1 mmol·L−1 pNA。混合体系加入96孔板中,在酶标仪上37 ℃反应45 min,每5 min测量1次吸光值[12]。
-
构建缺失突变体:使用pipA和pipB deletion引物以Xcc 8004基因组为模板扩增pipA和pipB基因上下游片段Hind III/BamH Ⅰ, BamH Ⅰ/EcoR Ⅰ酶切上下游片段,使用T4连接酶连接到Hind Ⅲ/EcoR Ⅰ酶切的pK18mobSacB载体中,获得敲除载体pK18-pipA和pK18-pipB。通过三亲接合法将重组载体转化至Xcc 8004中,经过两步同源重组方法获得突变体菌株ΔpipA、ΔpipB[13]。
基因互补:以Xcc
8004 基因组为模板,扩增pipA、pipB基因片段,Hind Ⅲ/EcoR Ⅰ酶切后克隆到相同酶切的广宿主载体pHMⅠ,获得互补载体pHMⅠ-pipA、pHMⅠ-pipB。三亲接合杂交法进行回补和交叉回补,获得相关菌株(WT、ΔpipA、ΔpipB、ΔpipA/ΔpipB、pipA/ΔpipA、pipB/ΔpipB、pipA/ΔpipB、pipB/ΔpipA、pipA/ΔpipA/ΔpipB、pipB/ΔpipA/ΔpipB)[13]。 -
待测菌株活化于NRSP(NYG:1% Tryptone; 1% Glycerol; 0.5% Yeast-extract+Rif+Sp)平板培养基中28 ℃,使用灭菌ddH2O将菌体清洗下来,将OD600值调至1.0,剪叶法接种45 d左右生长一致、长势良好的‘京丰一号’甘蓝,每种菌株至少接种10片叶片,从0 d开始,每5 d进行一次观测,待病斑长度有显著差异时对其进行拍照并测量病斑长度[14]。
-
将相关待测菌株在NRSP固体培养基上活化,并接种于NYG液体培养基中,28 ℃,160 r·min−1振荡培养至OD600值约为1.0,将菌液进行10~10−4梯度稀释,点接在NRSP培养基上,48 h后观察其生长状况;将菌液以1∶100(体积比)转接至新鲜的NRSP液体培养基中,间隔一定时间测量菌液浓度,绘制细菌生长曲线。
-
在NYG培养基中,以28 ℃和180 r·min−1的条件培养待测菌株,直至OD600约为1.0,用无菌ddH2O清洗2次,重悬于ddH2O中,使OD600值为1.0。取2 µL不同待测菌液分别接种到含有0.5%羧甲基纤维素、0.1%可溶性淀粉或2%脱脂奶粉的NRSP固体培养基上。28 ℃倒置培养至菌圈直径为1 cm左右。分别染色:纤维素平板:0.1%刚果红染色30 min,加入1 mol·L−1 NaCl于水平摇床60 r·min−1清洗1~2 min,测量菌圈直径及水解圈直径(cm)。淀粉平板:1∶100的I2/KI (0.08 mol·L−1 I2,3.2 mol·L−1 KI)染色1~10 min,测量菌圈直径和水解圈直径(cm)。脱脂奶粉平板:直接观察,测量菌圈直径和水解圈直径(cm)。水解能力以水解圈直径与菌圈直径之差表示(cm)[15]。
-
在NYG培养基中,以28 ℃和180 r·min−1的条件培养待测菌株,直至OD600值约为1.0,用无菌ddH2O清洗2次,重悬于ddH2O中,使OD600值为1.0。无菌牙签蘸取菌液5 s后,垂直接种于游动培养基 (0.03% Tryptone, 0.03% Yeast-extract, 0.25% Agar) 中。28 ℃正置培养至可观察到显著游动圈时,测量菌体游动圈的直径(cm)。蠕动性:取上述稀释菌液2 µL等间距点接到蠕动性培养基(1% Tryptone, 0.5% Yeast-extract, 2% Glucose, 0.6% Agar)上,28 ℃正置培养至可观察到显著蠕动圈时,测量菌圈的直径(cm)[16]。
-
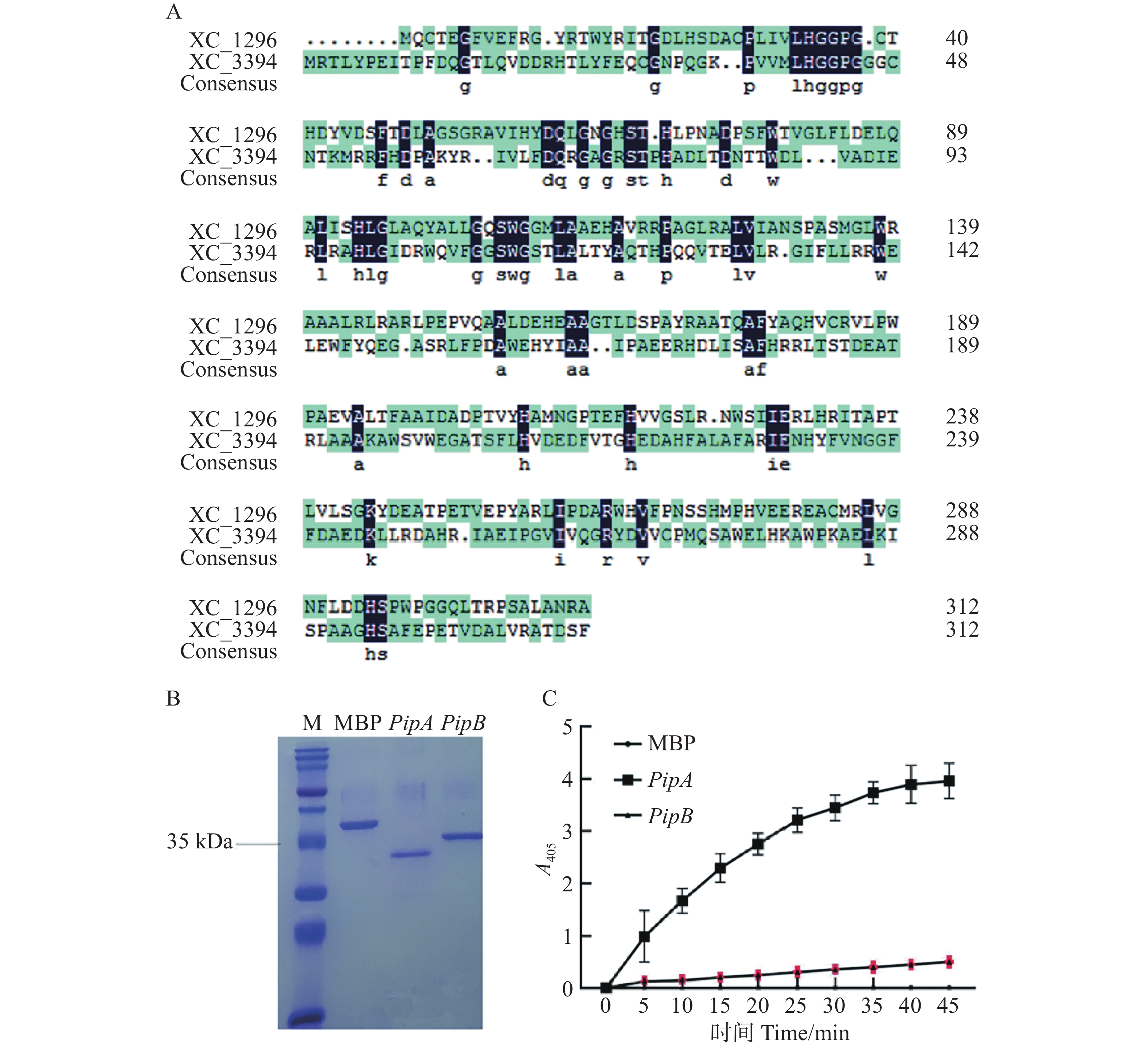
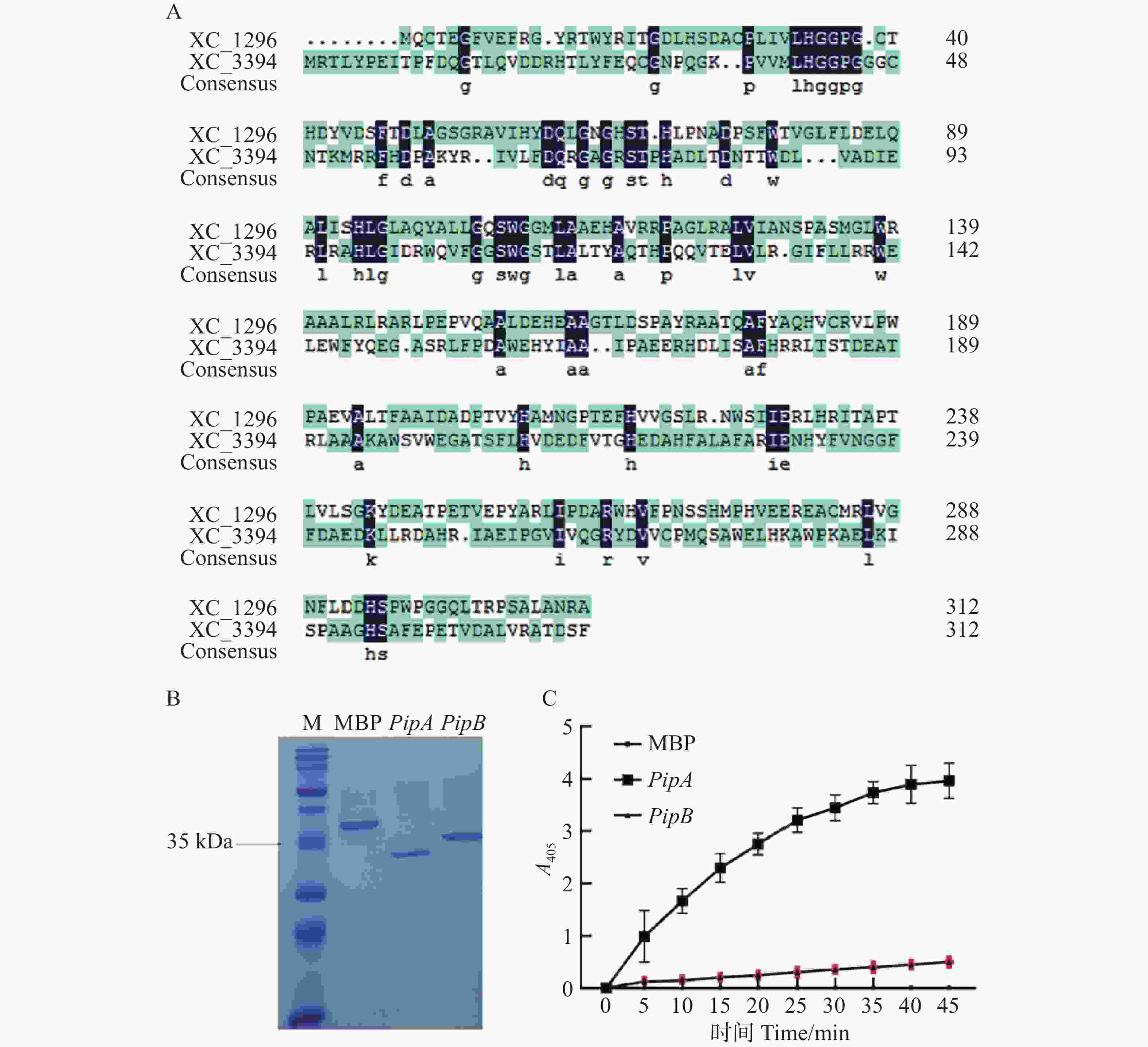
虽然PipA已被证实为PIPase,但在Xcc中是否还有相同活性的蛋白并不清楚。为此,通过同源搜索发现,Xcc中还有1个与PipA同源的蛋白(XC3394),但其具体功能还未被鉴定。对二者进行了序列比对,其核酸序列相似性为44.72%、氨基酸序列相似性为16.46%(图1−A)。为分析XC3394是否具有PIPase活性,本研究在大肠杆菌中表达和纯化了PipA和XC3394(图1−B),利用pNA为底物测定PIPase活性。如图1−C所示,PipA和XC3394均能分解p-NA。虽然XC3394分解p-NA的能力低于PipA,但其仍具有PIPase活性。因此,将XC3394命名为PipB。
-
虽然已有研究表明PipA调控Xcc致病性[17],但PipB的功能并不清楚。为分析PipB是否调控Xcc致病力,构建了pipA和pipB的突变体及互补菌株(WT、ΔpipA、ΔpipB、ΔpipA/ΔpipB、pipA/ΔpipA、pipBΔpipB、pipA/ΔpipB、pipB/ΔpipA、pipA/ΔpipA/ΔpipB、pipB/ΔpipA/ΔpipB),通过剪叶法接种检测了这些菌株的致病力。结果显示(图2),与野生型相比,pipA突变菌株致病力极显著降低,互补pipA后,部分恢复了致病力;pipB突变后致病性也显著降低;互补pipB恢复了突变体的致病力。互补pipA能够恢复pipB及pipA/pipB突变体的致病力,但互补pipB不能恢复pipA及pipA/pipB突变体的致病力。以上结果说明,PipA和PipB正调控Xcc致病力,但PipA和PipB功能不冗余,都调控Xcc的致病性。
-
为分析PipA和PipB调控Xcc致病性是否与细菌生长相关,比较了pipA和pipB的突变体及互补菌株(WT、ΔpipA、ΔpipB、ΔpipA/ΔpipB、pipA/ΔpipA、pipB/ΔpipB、pipA/ΔpipB、pipB/ΔpipA、pipA/ΔpipA/ΔpipB、pipB/ΔpipA/ΔpipB)的生长速度。如图3所示,无论是在液体还是固体培养基中,所有菌株生长速度都没有差异。因此,PipA和PipB可能不通过调控病原菌的生长来影响其致病力。
-
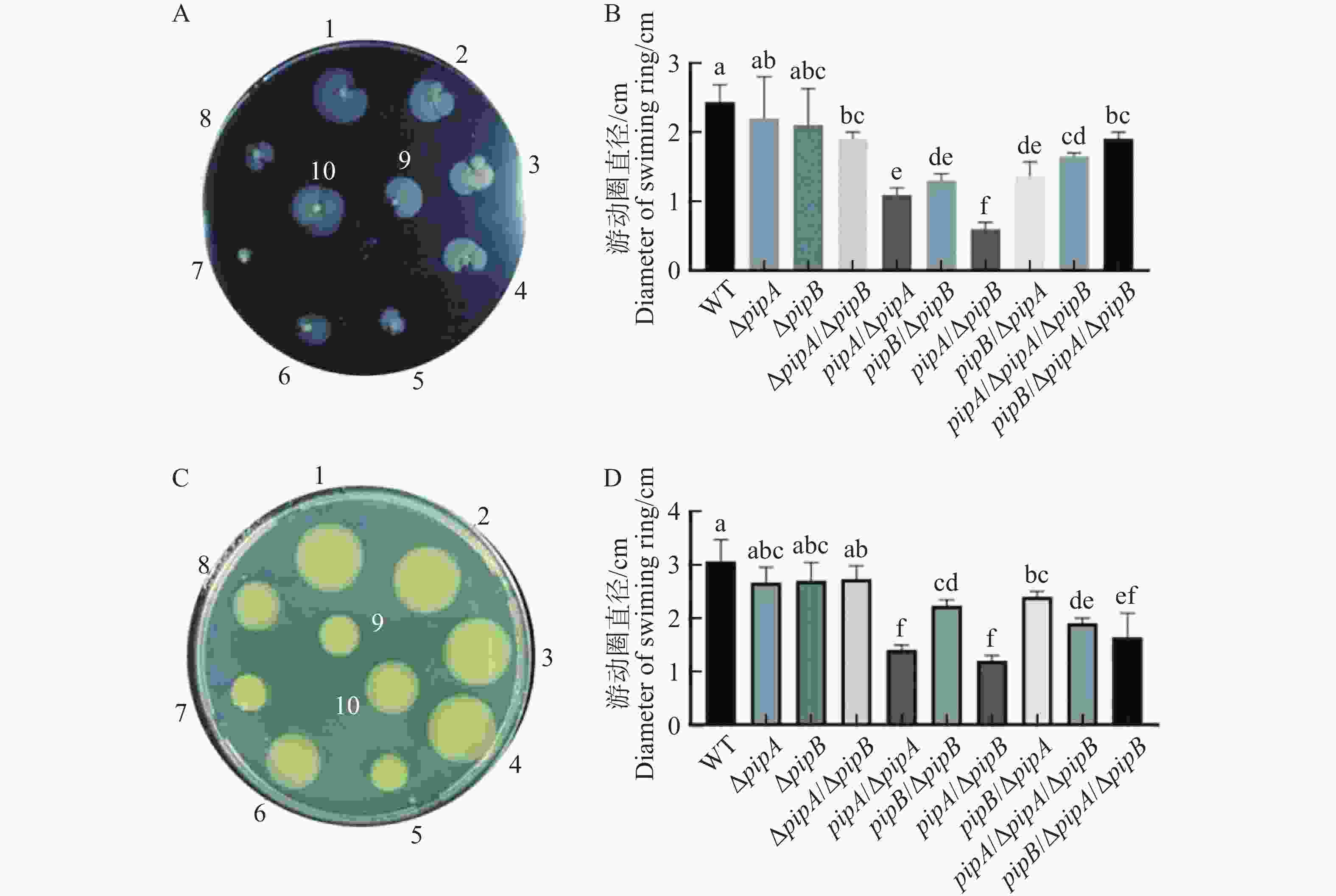
病原菌的运动性与其致病性相关,运动性在致病初期具有重要作用。细菌普遍的运动方式是游动和蠕动,均调控病原菌的致病力[18 − 19]。为此,本研究检测了pipA和pipB突变对Xcc运动能力的影响。细菌游动性和蠕动性实验发现pipA和pipB突变并不显著影响病原菌的运动性,但pipA和pipB回补菌株的运动性显著低于野生型及突变体菌株(图4)。该结果说明pipA和pipB表达量升高会显著抑制病原菌的运动能力。因此,在病原菌侵染植物过程中,pipA和pipB表达量升高可能导致病原菌运动能力降低,进而促进病原菌在维管束中的定殖。
-
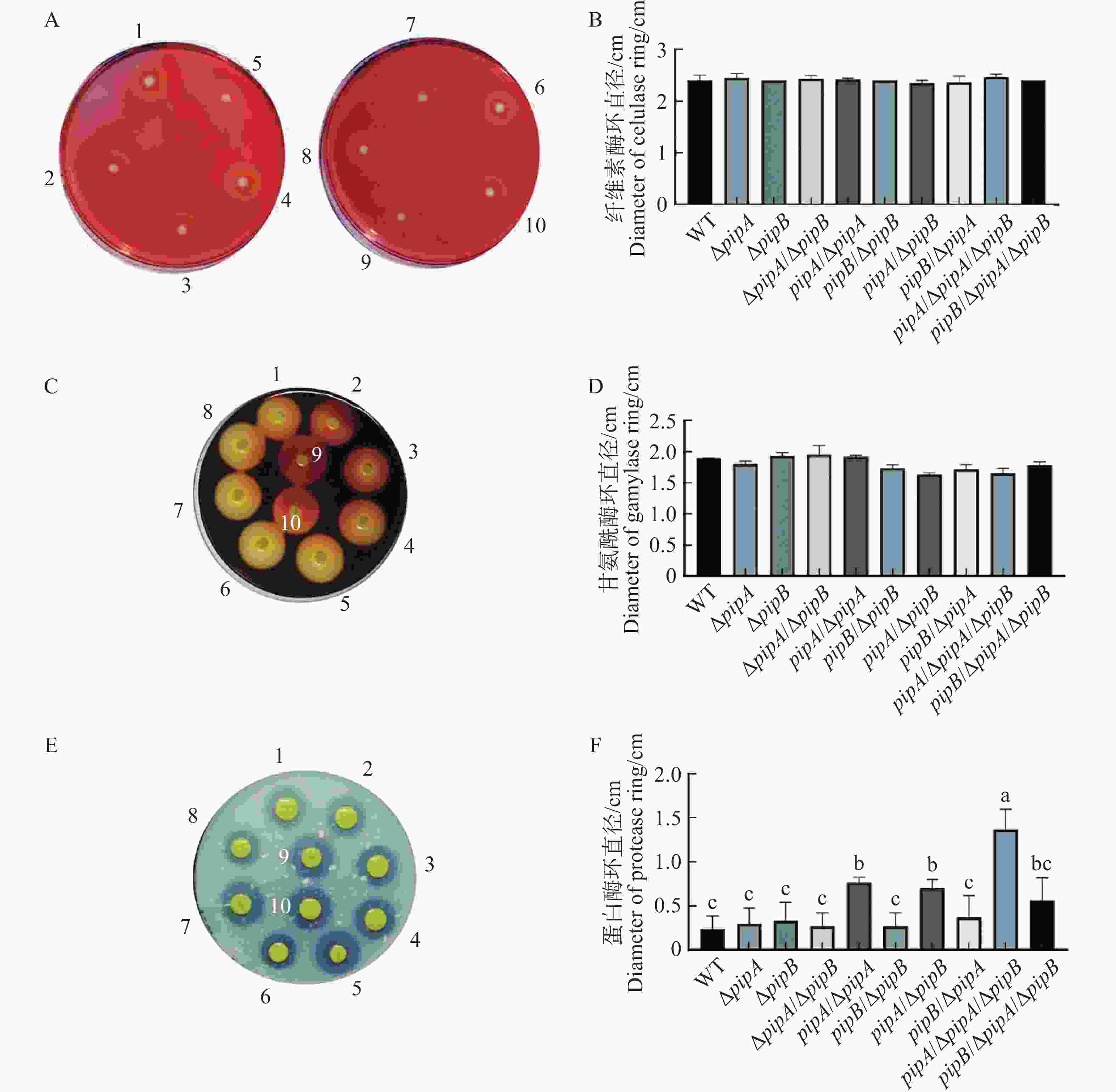
植物细胞壁是其抵抗病原菌侵染的一道屏障,病原菌分泌的多种细胞壁降解酶促进其入侵[20]。通过比较PIPase相关菌株各种胞外酶活性发现,pipA过表达使胞外蛋白酶活性增强,但不影响胞外纤维素酶和淀粉酶的变化,pipB突变及过表达均不影响Xcc胞外酶活性(图5)。因此,PipA可能通过调控胞外蛋白酶活性或分泌影响Xcc的致病力。
-
胞外多糖(Extracellular polysaccharide, EPS)可抑制防御反应来增强寄主植物的敏感性[21]。此外,EPS还可以包裹细菌以阻止宿主识别并使其在宿主组织中高效定殖[22]。为探究PipA和PipB是否参与胞外多糖的形成,通过将待测菌株接种在含有2%葡萄糖的NYG固体培养基上观察其菌落形态,以分析其FPS合成能力。结果显示(图6),所有菌株的菌落形态与野生型相比几乎没有差别。因此,PipA和PipB不调控胞外多糖的形成。
-
本研究旨在探究PipA和PipB是否具有PIPase活性,是否调控Xcc致病力以及是否存在功能冗余。体外酶活实验证实,PipB同PipA一样具有PIPase活性,暗示这2个PIPase可能存在功能冗余或特异性。已知PIPase在调控细菌的运动性、胞外酶合成等方面发挥了重要作用[9]。例如,在Xcc中,pipA基因表达受到上游群体感应调节因子XccR诱导,是Xcc致病力的重要调节因子,可抑制植物水杨酸信号通路,进而抑制植物免疫反应,促进病原菌侵染[9,23],但具体调控机制还不清楚。本研究发现,除PipA外,PipB同样是Xcc致病力的重要调控因子,表明2个PIPase均在Xcc与植物互作中起着非常重要的作用。
为探究PipA和PipB通过何种机制调控Xcc的致病力,本研究从细菌的生长、胞外酶活性和运动性等方面进行了研究。pipA和pipB突变和过表达对细菌生长、胞外纤维素酶、胞外淀粉酶活性和胞外多糖的产生均无显著影响,但对运动性有一定的影响,暗示PIPase可能通过调节Xcc运动性,进而影响其致病力。除运动性外,pipA的过表达还影响胞外蛋白酶活性。此外,PipA对运动性、蛋白酶活的影响远强于pipB。因此,PipA可能是Xcc主要的PIPase,在其致病力调控中起到主要作用,而PipB可能作为辅助性PIPase。但是交叉互补实验发现pipB并不能互补pipA突变导致的功能变化,其单独突变也能显著降低病原菌的致病力。以上结果表明PipA和PipB功能并不冗余,可通过调控不同致病相关途径来调控病原菌的致病力,但PipA和PipB调控Xcc致病力的具体机制还需进一步研究。
脯氨酸亚氨基肽酶调控野油菜黄单胞菌的致病力
DOI: 10.15886/j.cnki.rdswxb.20240079
 CSTR: 32425.14.j.cnki.rdswxb.20240079
CSTR: 32425.14.j.cnki.rdswxb.20240079
Proline imino-peptidases regulate the virulence of Xanthomonas campestris
-
摘要: 为探究脯氨酸亚氨基肽酶是否调控野油菜黄单胞菌野油菜致病变种(Xanthomonas campestris pv. campestris,Xcc)的致病力,首先通过生物信息学分析Xcc中潜在的脯氨酸亚氨基肽酶,然后在大肠杆菌中表达、纯化这些蛋白,体外检测其脯氨酸亚氨基肽酶酶活性,最后在Xcc中突变及回补这些基因,通过分析突变及回补菌株的致病力、生长特性、运动力和胞外酶活性,初步确定脯氨酸亚氨基肽酶在调控Xcc致病力中的作用。结果表明,Xcc编码两个脯氨酸亚氨基肽酶(PipA和PipB),突变其中任何一个都导致Xcc致病力下降。进一步分析发现pipA和pipB过表达均导致病原菌运动力增强。此外,pipA过表达提高了Xcc胞外蛋白酶活性,但pipB不具备这种活性。因此,脯氨酸亚氨基肽酶PipA和PipB可能通过调控Xcc的运动性,进而影响病原菌的致病能力,PipA还可能通过调控胞外蛋白酶活性,进而影响Xcc致病力。Abstract: To investigate if the proline aminopeptidases (PIPases) regulate the virulence of Xanthomonas campestris pv. campestris (Xcc), we first analyzed the putative PIPases in Xcc by bioinformatics; then expressed and purified these proteins, and detected their PIPase activities in vitro; finally studied the roles of these genes in Xcc virulence by analyzing the infective ability, growth rates, motility, and extracellular enzyme activities of the mutants and the coressponding complementary strains of these mutatns. The results indicate that Xcc encodes two PIPases, PipA and PipB. Mutation of either gene led to a decrease in Xcc virulence. Overexpression of pipA or pipB resulted in enhanced motility. In addition, pipA overexpression increased the activities of the extracellular proteases in Xcc, but pipB did not have this activity. Therefore, PipA and PipB may affect Xcc virulence by regulating its motility, and PipA may also regulate extracellular protease activity to affect Xcc infection.
-
Key words:
- Xanthomonas campestris pv. campestris /
- PIPase /
- Virulence /
- Motility
-
图 5 PipA和PipB对Xcc胞外酶活性的调控作用
平板法检测胞外纤维素酶(A)、淀粉酶(C)和蛋白酶(E)活性;B、D和F为A、C和E图的消化圈直径的统计结果;水解能力以水解圈直径与菌圈直径之差表示(cm)。
Fig. 5 The activities of extracellular enzymes are regulated by PipA and PipB
The activities of cellulase (A), amylase (C) and protease (E) analyzed by plate assays; The digestive diameters were also shown in B, D and F for cellulase, amylase and protease activities, respectively. The hydrolysis activity was shown as the difference between the diameter of the hydrolysis circle and the diameter of the colony circle (cm).
表 1 菌株和质粒来源
Table 1 Strains and plasmids used in this study
名称Name 相关特性Correlation characteristic 来源Source Strains Xcc 8004 A wild-type Xcc strain, a laboratory strain with spontaneous rifampicin-resistance Lab collections DH5α Genetically engineered recipient bacteria. Biomed BL21(DE3) Bacteria for protein expression Biomed ∆pipA The pipA deletion mutant of WT This study ∆pipB The pipB deletion mutant of WT This study ∆pipA/∆pipB The pipA and pipB deletion mutant of WT This study pipA/∆pipA The pipA complementation strain of ∆pipA This study pipB/∆pipB The pipB complementation strain of ∆pipB This study pipA/∆pipB The pipA complementation strain of ∆pipB This study pipB/∆pipA The pipB complementation strain of ∆pipA This study pipA/∆pipA/∆pipB The pipA complementation strain of ∆pipA/∆pipB This study pipB/∆pipA/∆pipB The pipB complementation strain of ∆pipA/∆pipB This study Plasmids pK18mobSacB Xanthomonas suicide vector, KanR [10] pHMI Broad host vector, SpR [11] pQE80L Protein expression vector, AmpR Qiagen pK18-pipA pK18mobSacB based plasmid for pipA deletion This study pK18-pipB pK18mobSacB based plasmid for pipB deletion This study pHMI-pipA pHMI based plasmid for pipA expression This study pHMI-pipB pHMI based plasmid for pipB expression This study pQE80L-pipA pQE80L based plasmid for pipA expression in E. coli This study pQE80L-pipB pQE80L based plasmid for pipB expression in E. coli This study 表 2 本研究所用引物
Table 2 Primers used in this study
引物Primer 序列Sequence(5′-3′) 用途Usages pipAddFF TCTGAAGCTTGCTGCTGGAGTTGTAGGAAG pipA deletion pipAddFR TCTGGGATCCGATGCGGTACCAGGTGCGAT pipAddRF ACGTGGATCCAACTTCCTCGACGACCACAG pipAddRR ACTCGAATTCAACGTAGTGCGCAGCACTAC pipBddFF TCTGAAGCTTGAGCGGTTCGATCTGATCGC pipB deletion pipBddFR TCTGGGATCCCATGCGTCGGTCCTGCGGGA pipBddRF ACGTGGATCCGACAGCTTCGCCTGAGAAGG pipBddRR ACTCGAATTCATCGGCTGCTGCAGGCTTTG pipAhmF ACCATGATTACGCCAAGCTTGATGCGCACGCTCTATCCCGA ΔpipA complementation pipAhmR GTAGAATTCTAGAGGGTACCAGACGGCACGATTGGCCAGCG pipBhmF ACCATGATTACGCCAAGCTTGATGCGCACGCTCTATCCCGA ΔpipB complementation pipBhmR GTAGAATTCTAGAGGGTACCAGGCGAAGCTGTCGGTCGCAC pipAqeF ACGTGGATCCATGCAGTGCACCGAGGGTTTC PipA expression pipAqeR ACGCAAGCTTAGACGGCACGATTGGCCAGCG pipBqeF ACGTGGATCCATGCGCACGCTCTATCCCGAG PipB expression pipBqeR ACGCAAGCTTAGGCGAAGCTGTCGGTCGCAC 注:下划线表示酶切位点。
Note: The enzyme cutting sites are underlined. -
[1] VICENTE J G, CONWAY J, ROBERTS S J, et al. Identification and origin of Xanthomonas campestris pv. campestris races and related pathovars[J]. Phytopathology, 2001, 91(5): 492 − 499. doi: 10.1094/PHYTO.2001.91.5.492 [2] 朱镜璇. 分子动力学模拟结合深度学习探究氨基酸突变或配体结合对酶活性的影响[D]. 长春: 吉林大学, 2021. [3] KAMARUDDIN S, AHMAD REDZUAN R, MINOR N, et al. Biochemical characterisation and structure determination of a novel cold-active proline iminopeptidase from the psychrophilic yeast, Glaciozyma antarctica PI12[J]. Catalysts, 2022, 12(7): 722. doi: 10.3390/catal12070722 [4] KUERMAN M, WANG R, ZHOU Y, et al. Metagenomic insights into bacterial communities and functional genes associated with texture characteristics of Kazakh artisanal fermented milk Ayran in Xinjiang, China[J]. Food Research International, 2023, 164: 112414. doi: 10.1016/j.foodres.2022.112414 [5] INTAD S. Biochemical activities of actinopain peptidases from actinomyces bacteria[D]. Hong Kong, China: The University of Hong Kong, 2023. [6] KAMARINOU C S, KIOUSI D E, REPANAS P, et al. Dissecting the genetic basis of the technological, functional, and safety characteristics of Lacticaseibacillus paracasei SRX10[J]. Microorganisms, 2024, 12(1): 93. doi: 10.3390/microorganisms12010093 [7] YAMAMOTO Y, USUKI H, IWABUCHI M, et al. Prolyl aminopeptidase from Streptomyces thermoluteus subsp. fuscus strain NBRC14270 and synthesis of proline-containing peptides by its S144C variant[J]. Applied and Environmental Microbiology, 2010, 76(18): 6180 − 6185. doi: 10.1128/AEM.01242-10 [8] YAMAMOTO Y, USUKI H, KUMAGAI Y, et al. Synthesis of prolyl-hydroxyproline using prolyl aminopeptidase from Streptomyces aureofaciens TH-3[J]. Process Biochemistry, 2011, 46(8): 1560 − 1564. doi: 10.1016/j.procbio.2011.04.009 [9] ZHANG L L, JIA Y T, WANG L, et al. A proline iminopeptidase gene upregulated in planta by a LuxR homologue is essential for pathogenicity of Xanthomonas campestris pv. campestris[J]. Molecular Microbiology, 2007, 65(1): 121 − 136. doi: 10.1111/j.1365-2958.2007.05775.x [10] SCHÄFER A, TAUCH A, JÄGER W, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum[J]. Gene, 1994, 145(1): 69 − 73. doi: 10.1016/0378-1119(94)90324-7 [11] HUYNH T V, DAHLBECK D, STASKAWICZ B J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity[J]. Science, 1989, 245(4924): 1374 − 1377. doi: 10.1126/science.2781284 [12] SUDO T, SHINOHARA K, DOHMAE N, et al. Isolation and characterization of the gene encoding an aminopeptidase involved in the selective toxicity of ascamycin toward Xanthomonas campestris pv. citri[J]. Biochemical Journal, 1996, 319(1): 99 − 102. doi: 10.1042/bj3190099 [13] BORN A, SOETBEER J, HENEN M A, et al. Ligand-specific conformational change drives interdomain allostery in Pin1[J]. Nature Communications, 2022, 13(1): 4546. doi: 10.1038/s41467-022-32340-x [14] 郭艺. Xcc suxB应答蔗糖信号调控机制的初步研究[D]. 海口: 海南大学, 2020. doi: 10.27073/d.cnki.ghadu.2020.001152 [15] 于燕燕, 夏影影, 吴可建, 等. XppI调控水稻白叶枯病菌致病力的潜在机制[J]. 热带生物学报, 2023, 14(6): 642 − 650. doi: 10.15886/j.cnki.rdswxb.20230024 [16] KOYANAGI T, HARA A, KOBAYASHI K, et al. Thermococcus sp. KS-1 PPIase as a fusion partner improving soluble production of aromatic amino acid decarboxylase[J]. AMB Express, 2021, 11(1): 178. doi: 10.1186/s13568-021-01340-3 [17] KAN J H, AN L, WU Y, et al. A dual role for proline iminopeptidase in the regulation of bacterial motility and host immunity[J]. Molecular Plant Pathology, 2018, 19(8): 2011 − 2024. doi: 10.1111/mpp.12677 [18] WADHWA N, BERG H C. Bacterial motility: machinery and mechanisms[J]. Nature Reviews Microbiology, 2022, 20(3): 161 − 173. doi: 10.1038/s41579-021-00626-4 [19] MOREIRA C G, WEINSHENKER D, SPERANDIO V. QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo[J]. Infection and Immunity, 2010, 78(3): 914 − 926. doi: 10.1128/IAI.01038-09 [20] 陈小云. 柑橘溃疡病菌致病相关基因生物学功能分析及XAC1992互作蛋白的初步筛选[D]. 广州: 华南农业大学, 2019. doi: 10.27152/d.cnki.ghanu.2019.001210 [21] YUN M H, TORRES P S, EL OIRDI M, et al. Xanthan induces plant susceptibility by suppressing callose deposition[J]. Plant Physiology, 2006, 141(1): 178 − 187. doi: 10.1104/pp.105.074542 [22] ALVAREZ A M. Black rot of crucifers[M]//SLUSARENKO A J, FRASER R S S, LOON L C. Mechanisms of resistance to plant diseases. Dordrecht: Springer, 2000: 21−52. [23] GHIFARI A S, TEIXEIRA P F, KMIEC B, et al. A mitochondrial prolyl aminopeptidase PAP2 releases N-terminal proline and regulates proline homeostasis during stress response[J]. The Plant Journal, 2020, 104(5): 1182 − 1194. doi: 10.1111/tpj.14987 -





 下载:
下载: