-
抗生素是由自然界中微生物分解代谢产生[1],并拥有在低浓度下抑制或杀死其他微生物的特征,目前被广泛应用于医疗领域来治疗感染性疾病[2]。抗生素被发现后,随之在环境和医疗领域被广泛应用,但这也导致病原微生物的耐药性不断增强,甚至出现了高度耐药性和强传染性的“超级细菌”,对全球公共卫生构成了严重威胁[3]。近年来,临床研究观察发现,抗生素在治疗常见细菌感染(如尿路感染、败血症、性传播感染和腹泻等疾病)时效果有所降低,导致感染死亡率上升[4]。2017 年,世界卫生组织发布了一份包含 12 种主要耐药细菌的清单,且多数病原菌对目前广泛使用的抗生素具有耐药性,如鲍曼不动杆菌(碳青霉烯耐药)、铜绿假单胞菌(碳青霉烯耐药)、肠杆菌科(碳青霉烯耐药、广谱β-内酰胺酶)、屎肠球菌(万古霉素耐药)、金黄色葡萄球菌(甲基霉素耐药)、幽门螺杆菌(克拉霉素耐药)、弯曲杆菌(氟喹诺酮耐药)、沙门氏菌(氟喹诺酮耐药)和淋病奈瑟菌(头孢菌耐药、氟喹诺酮耐药)、肺炎链球菌(青霉素不敏感)、流感嗜血杆菌(氨苄西林耐药)和志贺氏菌(对氟喹诺酮类药物产生耐药性)。其中,6 种病原菌(大肠杆菌、鲍曼不动杆菌、金黄色葡萄球菌、肺炎克雷伯菌、铜绿假单胞菌和肠杆菌)是日常生产生活中常见的致病菌[5]。
大肠杆菌(Escherichia coli,E.coli)是革兰氏阴性棒状杆菌,属于肠杆菌科,是常见的肠道细菌之一,具有鞭毛,能进行趋化性运动[6]。大肠杆菌广泛分布于人体和动物肠道中[7],是正常肠道菌群中重要的代表菌种[8]。然而,一些病原性大肠杆菌菌株能够导致肠道感染、尿路感染、败血症等多种临床疾病[9]。近年来,随着大肠杆菌快速获得多重耐药性,临床抗菌治疗正面临日益严峻的挑战[10]。
目前,药敏检测除了传统的基于培养基的纸片扩散法、肉汤微量稀释法外,还有基于荧光光电检测技术的PCR荧光杂交法、质谱法、流式细胞荧光法、拉曼光谱法等。这些方法存在着预处理复杂、操作繁琐、成本高、检测周期长、通量低等缺点,且只能定性,无法进行定量检测。面对病原微生物耐药性不断增加的严峻形势,新型抗微生物药物的研发已经陷入困境,特别是针对高度耐药的革兰氏阴性细菌,当前临床管线中的药物主要还是已知抗生素的衍生品[11]。因此,拥有高效快速的抗生素MIC检测工具和微生物耐药的生长动态监测平台,研发快速、高通量的新型药敏检测技术,对开发新型抗菌药物、控制细菌耐药性蔓延、优化抗菌治疗、维护公共卫生安全至关重要。
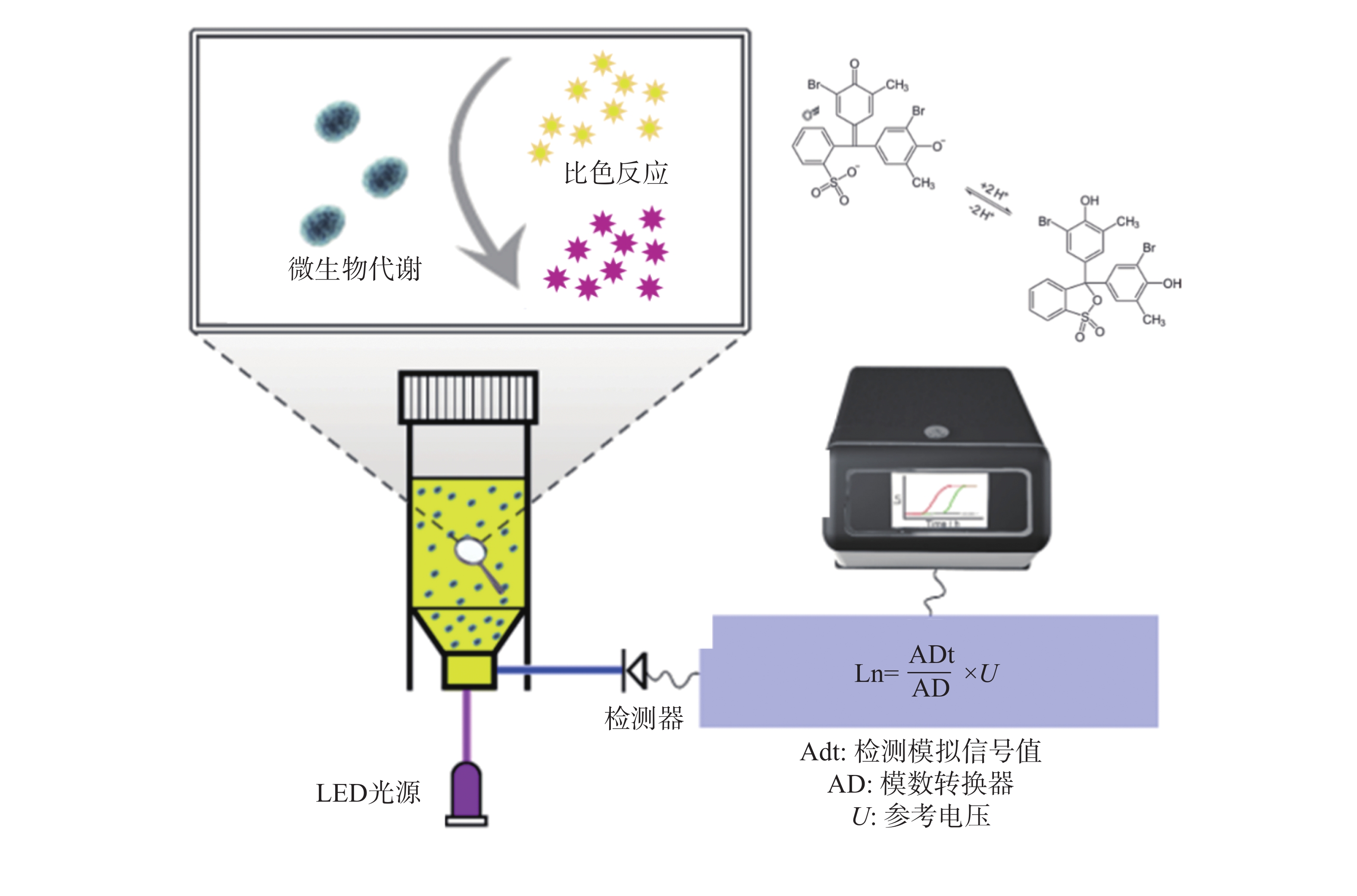
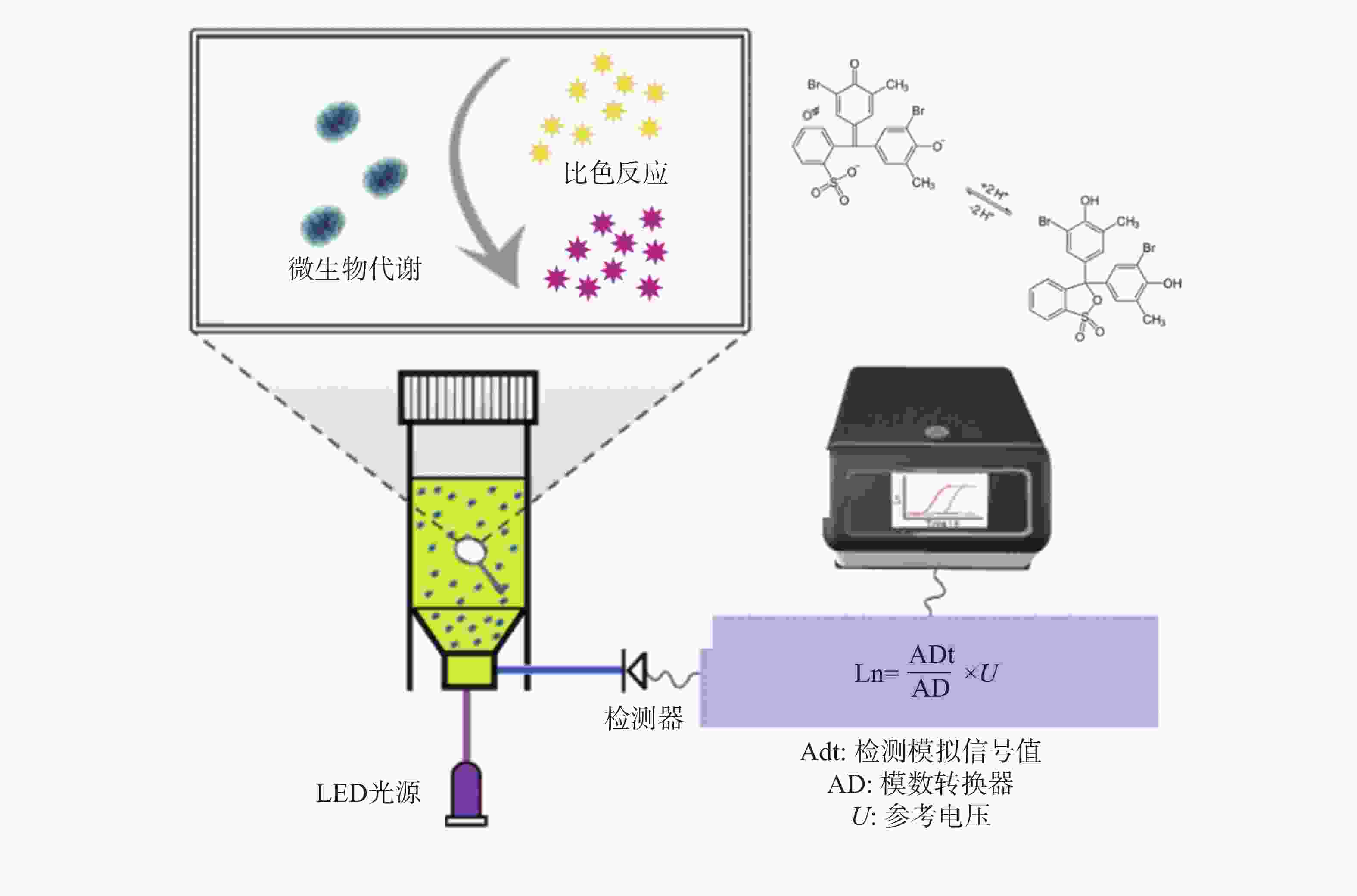
鉴于此,本研究团队开发了荧光光电微生物检测技术(图1),集成了光电传感、荧光探针和自动化培养, 能够高效、实时地监测微生物的生长及其耐药性状况。该系统由基于不同硬件功能的4个模块组成,包括主控制模块、光源模块、温度控制模块和检测模块。融合了选择性培养基培养技术、荧光光学检测技术、智能传感技术等,实现了微生物的自动判定或定量分析。本仪器的检测结果与国标的药敏检测法结果一致,证明了本技术的可靠性和准确性。本研究的成果将为临床抗生素治疗提供有益的指导,为未来耐药性管理和新药开发提供科学依据,为维护公共健康和医学进步作出贡献。
-
标准菌株为大肠杆菌CICC 10411购自中国工业微生物菌种保藏中心。将−20 ℃冷冻保存的菌液,于室温缓慢解冻,按 1∶100 的体积比例接种于无菌 LB 培养基中活化。
-
蛋白胨(Solarbio),胰蛋白胨(Oxoid),牛肉浸粉(Solarbio),氢氧化钠(西陇科学),葡萄糖(西陇科学),溴甲酚紫(生工生物),氯化钠(西陇科学),酵母粉(Oxoid)。氨苄青霉素(Solarbio),环丙沙星(Solarbio),萘啶酮酸(Alfa Aeser),庆大霉素硫酸盐(Solarbio),羧苄青霉素(Solarbio),头孢噻肟(Solarbio),氯霉素(源叶),多黏菌素 E(源叶)。萘啶酸药敏纸片(Liofilchem),环丙沙星药敏纸片(Liofilchem),多黏菌素E药敏纸片(Liofilchem),氨苄西林药敏纸片(Liofilchem)。
-
荧光光电微生物检测仪(微氪生物),PCR仪(伯乐),高速离心机(艾本德),低速离心机(上海卢湘仪),HVM1涡旋振荡器(珂淮仪器),电泳仪(北京六一),双人单面净化工作台(苏州净化),冷冻干燥机(宁波新芝生物),电磁炉(广东格兰仕),电热鼓风干燥箱(上海博迅),电热恒温培养箱(天津市赛得利斯),恒温培养振荡器(上海智城),立式压力蒸汽灭菌器(上海博迅医疗)。
-
用无菌生理盐水(v/v=0.85% NaCl)冲洗平板上菌落,混匀并调节至 0.5 麦氏浊度作为工作菌液。使用无菌棉拭子将菌液涂抹于麦康凯培养基平板表面。将药敏纸片贴附于平板边缘 15 mm 处,片间距离不低于24 mm,每平板贴 4~6 张纸片。贴片完成后倒置平板于恒温培养箱中,培养温度37 ℃,24~48 h后观测抑菌圈直径。
-
将10 g 蛋白胨、1.5 g 牛肉浸粉、0.042 g 氢氧化钠、5 g 葡萄糖、2.5 g NaCl、0.02 g 溴甲酚紫添加溶解到1 L超纯水中制成菌落总数培养基。使用 NaOH(1 mol·L−1)调节 pH 至 7.2 左右,灭菌后待用。 从对数生长期的原始大肠杆菌菌液中取 50 μL,用 LB培养基调整至 OD600=0.5。分别向检测管中加入1 mL 菌液和4 mL含有不同浓度抗生素的培养基,混匀。阳性对照(只加菌液),阴性对照(加生理盐水),设置合适的培养温度及培养时间。
-
从对数生长期的原始大肠杆菌菌液中取 50 μL,用 LB培养基调整至 OD600=0.5 。制备工作液浓度为0×、0.25×、0.5×、1×、2×MIC浓度的萘啶酮酸、羧苄青霉素溶液。吸取 1 μL 含药菌液加入荧光光电微生物检测仪试剂中,每个处理重复3次。放置于荧光光电微生物检测仪中,设置 24 h、37 ℃ 监测微生物生长情况。当微生物处于对数生长期时添加不同浓度的抗生素。设定荧光光电微生物检测仪每5 min 连续监测耐药菌浓度变化,生长进入平台期后停止检测。以OD值为纵坐标,时间为横坐标,导出时间动力学杀伤曲线图。
-
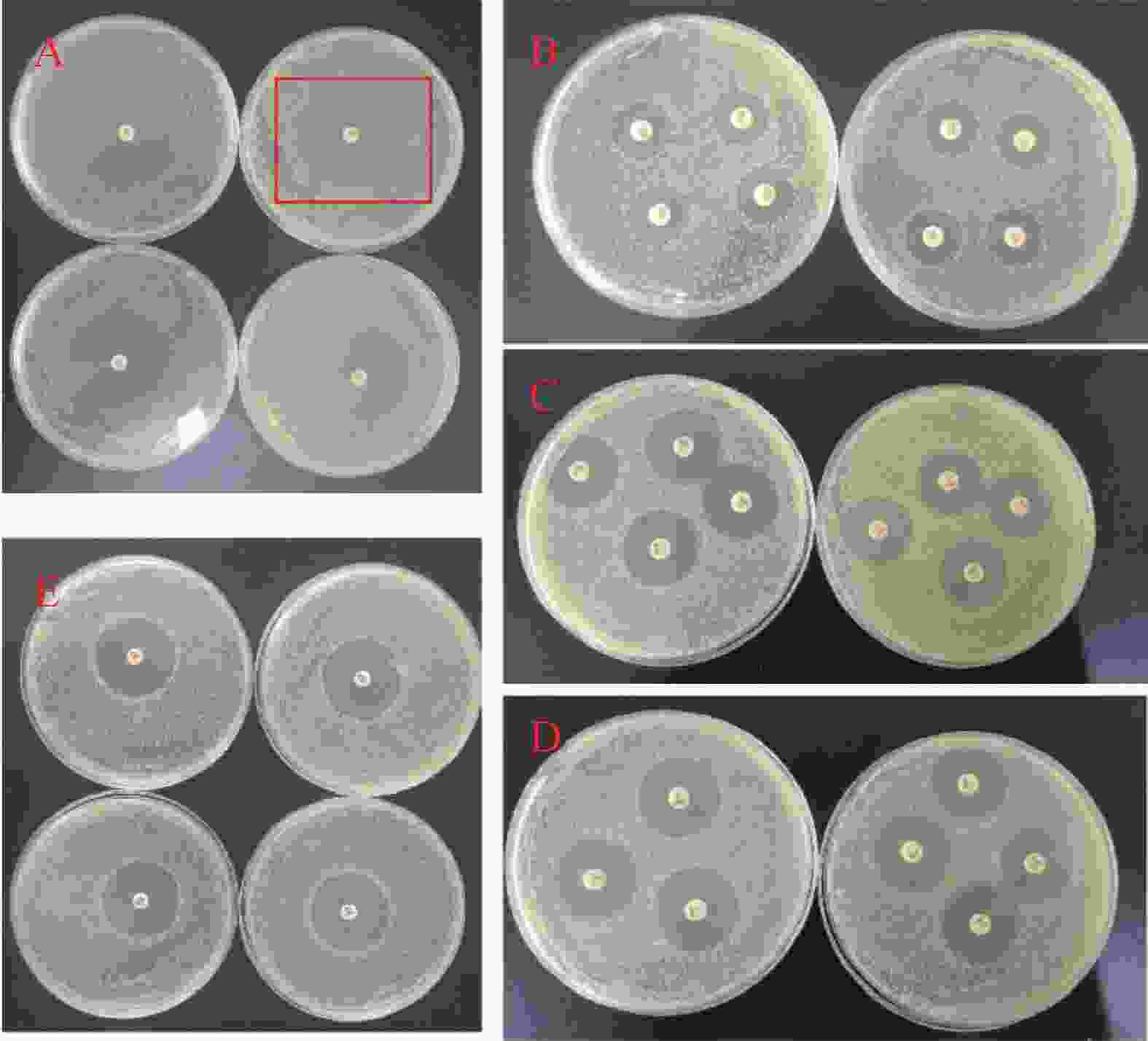
环丙沙星(CIP)属于四环素类喹诺酮广谱抗生素,作用于细菌DNA合成过程[12],实验中5 μg 的药敏纸片形成平均2 cm 直径的抑菌环。萘啶酮酸(NA)属于萘酚酮类抗生素[13],主要通过抑制细菌DNA合成发挥抗菌作用[14],30 μg 下形成 1.8 cm直径的抑菌环。氨苄青霉素(AMP)属于β-内酰胺类抗生素,能抑制细菌细胞壁的最后一步交联反应[15],10 μg 药敏纸片产生的 1.5 cm 直径的抑菌环。硫酸黏菌素(CS)又称多黏菌素 E,属于糖肽类抗生素,能够溶解细菌的细胞膜而发挥杀菌作用[16],10 μg 浓度的药敏纸片仅形成 1.1 cm 直径的抑菌环(图2)。
将这些数据进行等量药物换算,得到:1 μg CIP抑菌环的直径为 0.4 cm,1 μg NA抑菌环的直径为 0.06 cm;1 μg AMP抑菌环的直径为 0.15 cm;1 μg CS抑菌环的直径为 0.11 cm。结果表明:环丙沙星、氨苄青霉素、多黏菌素E、萘啶酮酸对大肠杆菌的抑菌效果由强至弱依次为环丙沙星>氨苄青霉素>多黏菌素E>萘啶酮酸。
-
大肠杆菌作为一种广泛存在的革兰氏阴性致病菌[17],其代谢活动会产生多种有机酸,导致周围环境pH下降,呈现酸性特征。溴甲酚紫(Bromothymol Blue, BTB)是一种常用的pH指示剂,能够根据溶液酸碱度呈现不同的颜色变化,在酸性条件下显现黄色,中性时为绿色,碱性环境下则为蓝色[18] (图3)。利用这一性质,可以将BTB引入大肠杆菌培养基中,通过荧光光电微生物检测仪内置的比色模型来确定菌液的浓度。
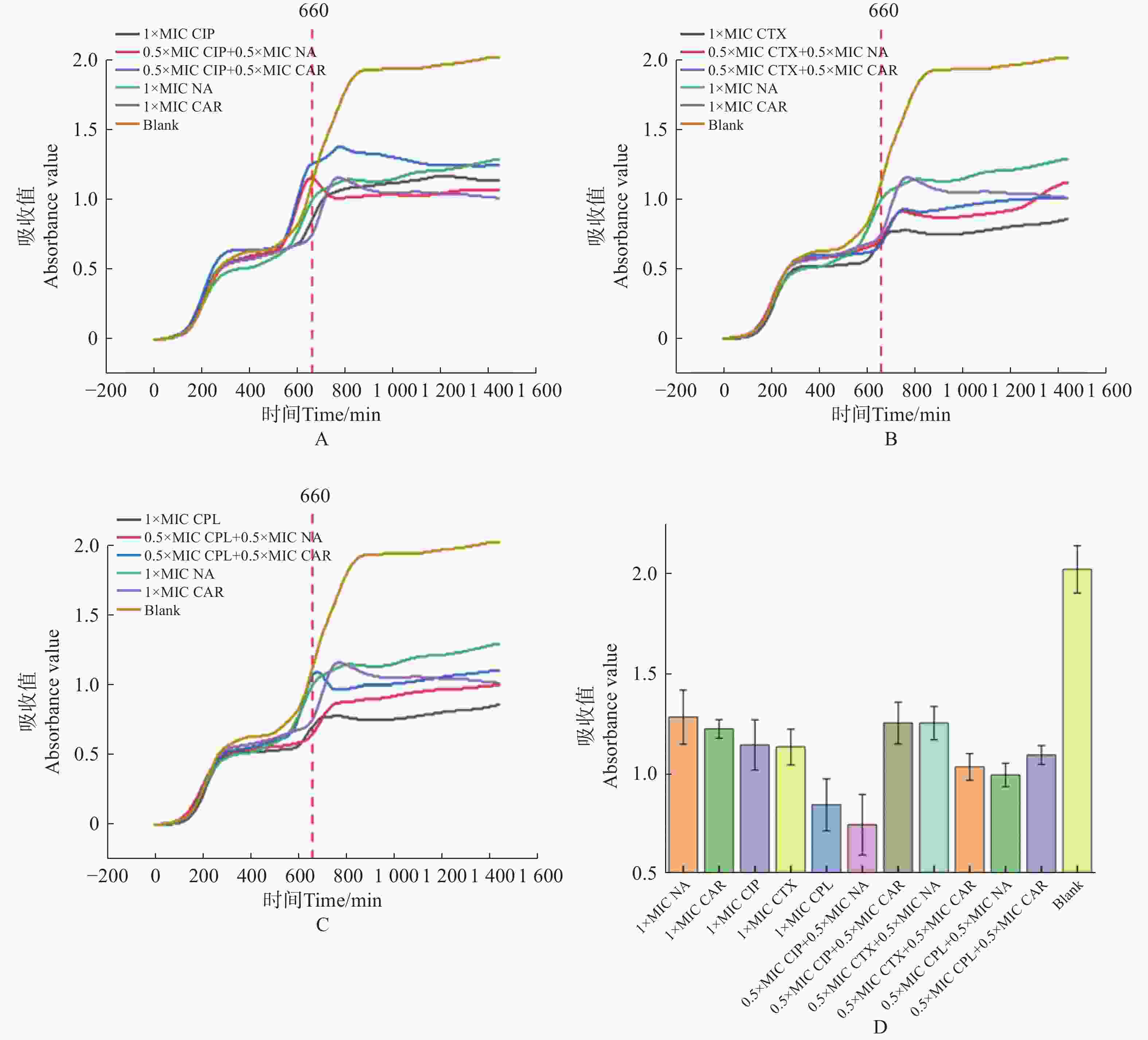
使用荧光光电微生物检测仪,检测不同抗生素对大肠杆菌的最小抑菌浓度(MIC)。如图4。
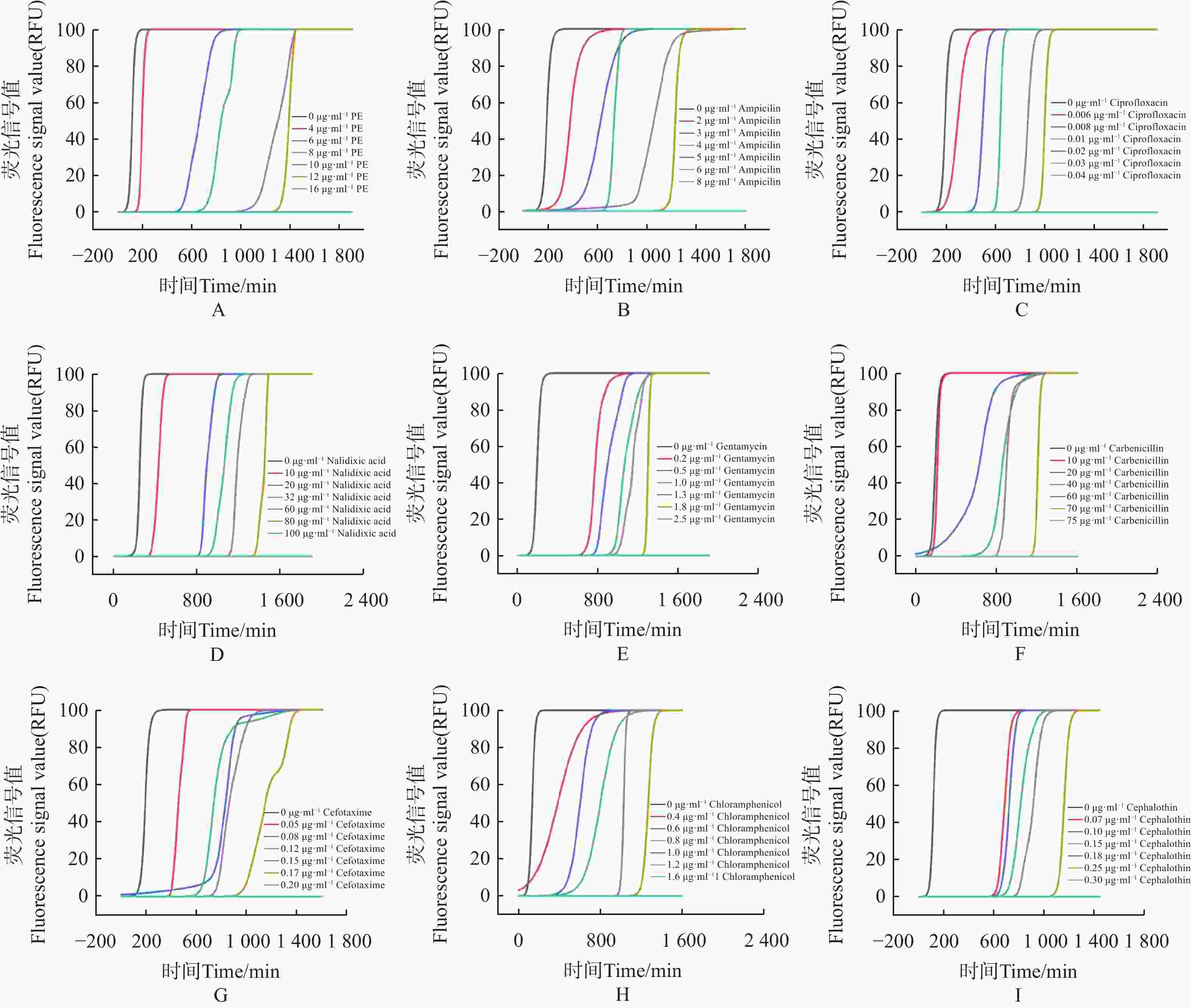
图 4 抗生素最低抑菌浓度检测
Figure 4. Detection of minimal inhibitory concentration of antibiotics against E.coli
1)多黏菌素 E(polymyxin E,PE),选择 0、4、6、8、10、12、16 μg·mL−17个质量浓度梯度,得到 PE 的MIC为16 μg·mL−1(图4-A)。
2)氨苄青霉素(ampicillin Capsules,AMP),选择0、2、3、4、5、6、8 μg·mL−17个质量浓度梯度,得到AMP的MIC为 8 μg·mL−1(图4−B)。
3)环丙沙星(ciprofloxacin,CIP),选择 0、0.006、0.008、0.01、0.02、0.03、0.04 μg·mL−17个质量浓度梯度,得出CIP的MIC为 0.04 μg·mL−1(图4−C)。
4)萘啶酮酸(nalidixic Acid,NA),选择 0、10、20、32、60、80、100 μg·mL−17个浓度梯度,得到 NA的MIC为 100 μg·mL−1(图4−D)。
5)庆大霉素(gentamicin,GM),选择0、0.2、 0.5、1、1.3、 1.8、2.5 μg·mL−17个质量浓度梯度,得到 GM的MIC为 2.5 μg·mL−1(图4−E)。
6)羧苄青霉素(carbenicillin,CAR),选择0、10、20、40、60、70、75 μg·mL−1 7个质量浓度梯度,得到CAR的MIC为75 μg·mL−1 (图4−F)。
7)头孢噻肟(cefotaxime,CTX),选择0、0.05、0.08、0.12、0.15、0.17、0.2 μg·mL−17个质量浓度梯度,得到 CTX 的MIC为 0.2 μg·mL−1(图4−G)。
8)氯霉素(chloramphenicol,CPL),选择0、0.4、0.6、0.8、1、1.2、1.6 μg·mL−1 7个质量浓度梯度,得到CPL的MIC为 1.6 μg·mL−1(图4−H)。
9)噻孢霉素(cephalothin,CET),选择 0、0.07、0.1、0.15、0.18、0.25、0.3 μg·mL−1 7个质量浓度梯度,得到 CET 的MIC为 0.3 μg·mL−1 (图4−I)。
结果表明,通过荧光光电微生物检测仪,得到不同抗生素对大肠杆菌的杀菌效果依次为环丙沙星 > 头孢噻肟 > 噻孢霉素 > 氯霉素 > 庆大霉素 > 氨苄青霉素 > 多黏菌素E > 羧苄青霉素 > 萘啶酮酸,证明了荧光光电微生物检测仪在抗生素耐药分析上等效于K-B 纸片法。
-
将获得的MIC数值和美国临床与实验室标准化研究所(CLSI)给出的临床药物断点值[19]对比。萘啶酮酸的MIC值为100 μg·mL−1(CLSI断点:敏感≤64 μg·mL−1),羧苄青霉素的MIC值为75 μg·mL−1(CLSI断点:敏感≤32 μg·mL−1),判断大肠杆菌对萘啶酮酸和羧苄青霉素耐药。分别配制0×MIC、0.25×MIC、0.5×MIC、1×MIC、2×MIC5个浓度梯度的萘啶酮酸和羧苄青霉素,进行动力学杀伤曲线测定。将E.coli预先培养11 h 后,加入不同浓度的萘啶酮酸和羧苄青霉素。结果表明,萘啶酮酸加入5 h 后(横坐标
1000 处),羧苄青霉素加入2 h后, E.coli 的生长呈现明显下降趋势,细菌的数量开始减少。2x MIC浓度的萘啶酮和羧苄青霉素,表现出明显的杀菌效果(图5−A,图5−B)。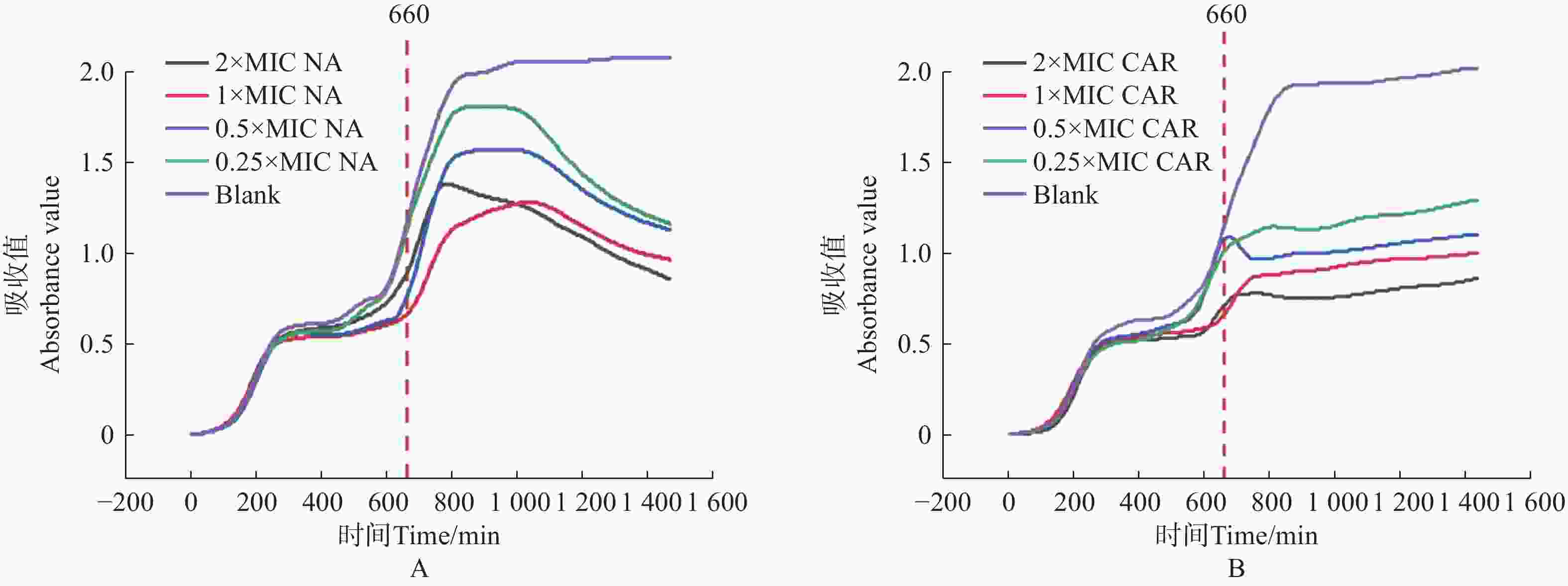
图 5 萘啶酮酸和羧苄青霉素对 E.coli 的动力学杀伤曲线
Figure 5. Kinetic killing curve of nalidixic acid and carbenicillin against E.coli
将萘啶酮酸和羧苄青霉素分别与环丙沙星、头孢噻肟、氯霉素联用,来确定对 E.coli 的协同杀菌效果。E.coli预先培养11 h 后,加入不同组合的抗生素。实验结果表明,0.5 倍 MIC 浓度的萘啶酮酸+0.5 倍 MIC 浓度的环丙沙星,对 E.coli 的杀伤效果最好(图6−A、图6−D)。0.5 倍 MIC 浓度的羧苄青霉素+0.5 倍 MIC 浓度的环丙沙星,对 E.coli杀菌效果最差(图6−A、图6−D)。
-
本研究基于传统药敏检测方法中存在的痛点,开发了荧光光电检测技术,实现了自动化培养、高通量检测、快速药物动力学杀伤曲线检测、快速抑菌浓度测定等。通过与K-B纸片法进行对比,证明了本方法的可靠性和准确性。通过本系统研究发现,0.5 × MIC 浓度环丙沙星 + 0.5× MIC 浓度萘啶酮酸对大肠杆菌起到很好的杀伤效果。环丙沙星抑制DNA解旋酶MCM2-7活性,抑制DNA的复制[20];同时能够抑制拓扑异构酶Ⅱ的活性,阻止线粒体DNA的复制起始[21];在结核杆菌体内能够抑制DNA的修复[22]。萘啶酮酸作为第一代喹诺酮类抗生素,能特异性抑制金黄色葡萄球菌的拓扑异构酶,抑制DNA的复制[23]。两种抗生素联用,可能协同干扰细菌DNA的合成,表现出协同增效的抗菌作用。羧苄青霉素作为β-内酰胺类抗生素[24],能够抑制细菌细胞壁的合成,具有广谱杀菌活性,能有效对抗革兰氏阴性和革兰氏阳性细菌[25]。本研究发现0.5×MIC浓度环丙沙星 + 0.5 × MIC浓度的羧苄青霉素对大肠杆菌的杀伤效果最差,这可能是由于β-内酰胺会诱导细菌高表达β-内酰胺酶,而环丙沙星作为喹诺酮类抗生素[26],会加剧细菌对喹诺酮类药物的耐药性,因此出现拮抗作用。
综上所述,荧光光电微生物检测仪可作为检测抗生素对微生物生长动力学影响的有效工具,能够快速甄别靶标菌株对某种抗生素的敏感性,为临床治疗和药敏实验提供了快速药敏检测平台。与传统微生物药敏检测相比,荧光光电微生物检测仪能够快速得出微生物的耐药情况。同时,弥补了传统微生物分离培养方法的不足,简化了多步骤、周期长的药敏实验过程,能够为医疗系统节省大量成本,缩短检测周期,提高患者的治愈率。
Drug sensitivity analysis of Escherichia coli based on fluorescence photoelectric detection
-
摘要: 本研究基于荧光光电微生物检测仪,研究不同抗生素对大肠杆菌的杀菌效果,结果证明使用荧光光电微生物检测仪进行微生物耐药分析的结果等效于K-B 纸片法药敏实验。相对于传统方法,荧光光电微生物检测仪能够快速鉴定临床标本中的微生物耐药情况,尤其对于苛养菌和其他生长缓慢的菌株,能够及时为抗生素的临床应用和病原菌的耐药性分析提供参考与借鉴。本研究使用 9 种抗生素对大肠杆菌进行耐药性检测,结果表明大肠杆菌对萘啶酮酸耐药性最高,最小抑菌浓度(minimal inhibitory concentration,MIC)为 100 μg·mL−1。大肠杆菌对头孢噻肟的耐药性最低,最小抑菌浓度为 0.2 μg·mL−1。将萘啶酮酸分别和环丙沙星、头孢噻肟、氯霉素联用,结果表明 0.5 倍 MIC 浓度的萘啶酮酸与 0.5 倍 MIC 浓度的环丙沙星联用,起到的杀菌效果较好。
-
关键词:
- 微生物耐药 /
- 大肠杆菌 /
- 荧光光电微生物检测仪 /
- 抗生素
Abstract: The bactericidal effects of different antibiotics on Escherichia coli were analyzed by using fluorescence photoelectric microbial detection devices. The results prove that the antimicrobial analysis using fluorescence photoelectric microbial detection devices is equivalent to the K-B disk diffusion method. Compared to the traditional methods, the fluorescence photoelectric microbial detection method can be used to quickly identify microbial resistance in clinical specimens, especially of fastidious bacteria and other slow-growing strains, providing timely reference for the clinical application of antibiotics and drug resistance analysis of pathogens. The detection of the drug resistance of E. coli to 9 antibiotics showed that E. coli had the highest resistance to nalidixic acid, with a minimal inhibitory concentration (MIC) of 100 μg.mL−1. E. coli had the lowest resistance to cefotaxime, with a minimal inhibitory concentration of 0.2 μg.mL−1. Combination of nalidixic acid with ciprofloxacin, cefotaxime, or chloramphenicol indicated that the combination of 0.5 MIC concentration of nalidixic acid with 0.5 MIC concentration of ciprofloxacin had the best bactericidal effect. -
-
[1] DE SIMEIS D, SERRA S. Actinomycetes: a never-ending source of bioactive compounds: an overview on antibiotics production[J]. Antibiotics, 2021, 10(5): 483. doi: 10.3390/antibiotics10050483 [2] KOHANSKI M A, DWYER D J, COLLINS J J. How antibiotics kill bacteria: from targets to networks[J]. Nature Reviews Microbiology, 2010, 8(6): 423 − 435. doi: 10.1038/nrmicro2333 [3] SERWECIŃSKA L. Antimicrobials and antibiotic-resistant bacteria: a risk to the environment and to public health[J]. Water, 2020, 12(12): 3313. doi: 10.3390/w12123313 [4] NEWMAN D. Old and modern antibiotic structures with potential for today's infections[J]. ADMET and DMPK, 2022, 10(2): 131 − 146. doi: 10.5599/admet.1272 [5] WILLYARD C. The drug-resistant bacteria that pose the greatest health threats[J]. Nature, 2017, 543(7643): 15. doi: 10.1038/nature.2017.21550 [6] SUMBANA J J. Phenotypic and molecular characterization of extraintestinal pathogenic Escherichia coli and other Gram-negative invasive bacteria in Mozambique[D]. Sassari: University of Sassari, 2020. [7] SAROWSKA J, FUTOMA-KOLOCH B, JAMA-KMIECIK A, et al. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports[J]. Gut Pathogens, 2019, 11(1): 10. doi: 10.1186/s13099-019-0290-0 [8] KAPER J B, NATARO J P, MOBLEY H L. Pathogenic Escherichia coli[J]. Nature Reviews Microbiology, 2004, 2(2): 123 − 140. doi: 10.1038/nrmicro818 [9] SMITH J L, FRATAMICO P M. Escherichia coli as a Pathogen[M]//DODD C E R, ALDSWORTH T, STEIN R A, et al. Foodborne diseases. Amsterdam: Elsevier, 2017: 189-208. doi: 10.1016/B978-0-12-385007-2.00007-3 [10] PAITAN Y. Current trends in antimicrobial resistance of Escherichia coli[J]. Current Topics in Microbiology and Immunology, 2018, 416: 181 − 211. doi: 10.1007/82_2018_110 [11] BEYER P, PAULIN S. The antibacterial research and development pipeline needs urgent solutions[J]. ACS Infectious Diseases, 2020, 6(6): 1289 − 1291. doi: 10.1021/acsinfecdis.0c00044 [12] MILLANAO A R, MORA A Y, VILLAGRA N A, et al. Biological effects of quinolones: a family of broad-spectrum antimicrobial agents[J]. Molecules, 2021, 26(23): 7153. doi: 10.3390/molecules26237153 [13] DU M J, ZHANG D, HOU Y L, et al. Combined 2D-QSAR, principal component analysis and sensitivity analysis studies on fluoroquinolones’ genotoxicity[J]. International Journal of Environmental Research, 2019, 16(21): 4156. doi: 10.3390/ijerph16214156 [14] AL BAHRY S, SIVAKUMAR N, AL-KHAMBASHI M. Effect of nalidixic acid on the morphology and protein expression of Pseudomonas aeruginosa[J]. Asian Pacific Journal of Tropical Medicine, 2012, 5(4): 265 − 269. doi: 10.1016/S1995-7645(12)60037-6 [15] BHATTACHARJEE M K. Antibiotics that inhibit cell wall synthesis[M]//BHATTACHARJEE M K. Chemistry of antibiotics and related drugs. Cham: Springer, 2022: 55-107. doi: 10.1007/978-3-031-07582-7_3 [16] ANDRADE F F, SILVA D, RODRIGUES A, et al. Colistin update on its mechanism of action and resistance, present and future challenges[J]. Microorganisms, 2020, 8(11): 1716. doi: 10.3390/microorganisms8111716 [17] BRAZ V S, MELCHIOR K, MOREIRA C G, et al. Escherichia coli as a multifaceted pathogenic and versatile bacterium[J]. Frontiers in Cellular and Infection Microbiology, 2020, 10: 548492. doi: 10.3389/fcimb.2020.548492 [18] STOCKER M K, SANSON M L, BERNARDES A A, et al. Acid–base sensor based on sol–gel encapsulation of bromothymol blue in silica: application for milk spoilage detection[J]. Journal of Sol-Gel Science and Technology, 2021, 98(3): 568 − 579. doi: 10.1007/s10971-021-05529-7 [19] WEINSTEIN M P, LEWIS J S. The clinical and laboratory standards institute subcommittee on antimicrobial susceptibility testing: background, organization, functions, and processes[J]. Journal of Clinical Microbiology, 2020, 58(3): e01864 − 19. doi: 10.1128/JCM.01864-19 [20] SIMON N, BOCHMAN M L, SEGUIN S, et al. Ciprofloxacin is an inhibitor of the Mcm2-7 replicative helicase[J]. Bioscience Reports, 2013, 33(5): e00072. doi: 10.1042/BSR20130083 [21] HANGAS A, AASUMETS K, KEKÄLÄINEN N J, et al. Ciprofloxacin impairs mitochondrial DNA replication initiation through inhibition of Topoisomerase 2[J]. Nucleic Acids Research, 2018, 46(18): 9625 − 9636. doi: 10.1093/nar/gky793 [22] O'SULLIVAN D M, HINDS J, BUTCHER P D, et al. Mycobacterium tuberculosis DNA repair in response to subinhibitory concentrations of ciprofloxacin[J]. Journal of Antimicrobial Chemotherapy, 2008, 62(6): 1199 − 1202. doi: 10.1093/jac/dkn387 [23] AHMED M, KELLEY S O. Enhancing the potency of nalidixic acid toward a bacterial DNA gyrase with conjugated peptides[J]. ACS Chemical Biology, 2017, 12(10): 2563 − 2569. doi: 10.1021/acschembio.7b00540 [24] THOPPIL A A, SHARMA R, KISHORE N. Complexation of β-lactam antibiotic drug carbenicillin to bovine serum albumin: energetics and conformational studies[J]. Biopolymers, 2008, 89(10): 831 − 840. doi: 10.1002/bip.21021 [25] LIMA L M, DA SILVA B N M, BARBOSA G, et al. β-lactam antibiotics: an overview from a medicinal chemistry perspective[J]. European Journal of Medicinal Chemistry, 2020, 208: 112829. doi: 10.1016/j.ejmech.2020.112829 [26] JACOBY G A, STRAHILEVITZ J, HOOPER D C, et al. Plasmid-mediated quinolone resistance[J]. Microbiology Spectrum, 2014, 2(5): 475-503. doi: 10.1128/microbiolspec.PLAS-0006-2013 -




 下载:
下载: