-
在自然环境中,植物总会遭受各种生物或非生物的胁迫,在长期的进化过程中,自然也会形成一定的防御机制。转录因子在植物响应逆境胁迫过程中具有重要功能,它们通过与多个基因启动子区域顺式作用元件的特异性结合参与植物的生长、发育和压力信号传递等过程[1-2]。乙烯响应因子(ERF,Ethylene Response Factor)是植物对生物和非生物胁迫响应反应中的主要调节因子[3-5]。转录因子识别特定的基序,并作为特定基因的激活或抑制因子发挥作用[6]。ERF转录因子含有57~70个氨基酸组成的ERF域[7-9],可以特异地与存在于植物细胞内的病原菌响应基因启动子中的GCC-box(GCC-box:AGCCGCC)结合,从而激活或者抑制ET,JA偶联的防御途径[8,10]。在许多病程相关基因启动子区都有GCC-box,如β−1,3−葡聚糖酶、几丁质酶和渗透蛋白,转录因子通过与GCC-box的结合直接或间接调节PR基因的表达,从而介导这些关键基因在植物响应生物胁迫中发挥作用[11]。木薯(Manihot esculenta)不仅是主要的粮食和经济作物,也是一种重要的再生能源作物[12-13]。广西、广东、海南、云南和福建是我国木薯的主要种植省份。由于种植省份的特殊地理位置,使木薯经常受到台风、海水倒灌使土地海盐量升高等的影响,极大地影响了木薯的产量。如时涛,李超萍等[14-15]的研究表明,低温则使得木薯低产甚至绝收,湿热则容易让木薯受到病害的威胁。目前,已发现ERF转录因子在木薯应对生物或非生物胁迫时能够发挥一定的作用[16]。因此,笔者利用生物信息学手段,挖掘木薯ERF转录因子的下游靶基因,旨在为木薯响应生物及非生物胁迫分子机理的解析提供基因资源和理论基础。
-
供试材料为华南8号(SC8, South China 8)品种木薯,由本实验室种植和保存。菌株:由本实验室在广西分离的Xam11菌株。主要试剂:多糖多酚植物总RNA提取试剂盒(TAINGEN, DP441),反转录试剂盒(Thermo,K1622), TB GREENII(TaKaRa,RR820A), DNA Marker(TaKaRa),引物由上海生工生物工程有限公司合成。
-
木薯ERF转录因子靶基因的生物信息学分析通过相应的在线数据库和软件完成。在Phytozome网站上下载木薯全基因组数据。利用在线网站Softberry对木薯全部基因的启动子区进行含有GCC-box顺式作用元件的基因的筛选;利用MapInspect软件分析MeBFs基因的染色体定位情况;在NCBI网站的SRA(Sequence Read Archive)数据库中下载病原菌侵染木薯的转录组数据。转录组数据来源于用Xam弱致病菌株ORST4和强致病菌株ORST4+TALE1侵染苗龄8周的木薯(品种为MCOL1522)组培苗的实验[17]。计算木薯ERF转录因子靶基因在这些转录组中的FPKM值,并将相互之间表达差异大于2倍的76个基因用Heatmap进行可视化;在Phytozome数据库中下载ERF转录因子靶基因ATG上游2 000 bp序列,利用PlantCARE在线网站对这2 000 bp序列的顺式作用元件进行预测分析,筛选与生物胁迫或非生物胁迫相关的顺式作用元件,再利用TBtools软件进行可视化分析。
-
用100 mg·L−1 ACC(1−氨基环丙烷−1−羧酸)喷施处理木薯(SC8)组培幼苗,处理后分别在0,0.5,1,2,4,6 h取样。用刀片刮伤法给苗龄30 d的木薯田间苗接种Xam11病原菌,分别在接种后0,5,8,24,48 h取样。取样时设置3个生物学重复,其中以10 μmol·L−1 MgCl2溶液作为病原菌处理的负对照,ddH2O作为激素处理的负对照。采取的叶片鲜样用液氮速冻,置于−80 ℃冰箱备用。
用多糖多酚植物总RNA提取试剂盒(TAINGEN,DP441)提取样品的RNA,用1.5%的琼脂糖凝胶电泳检测提取的总RNA的质量。以获得的总RNA为模板,利用反转录试剂盒(Thermo,K1622)进行cDNA反转录。反转录完成后,用木薯内参基因EF1a进行RT-PCR检测cDNA,以cDNA为模板进行后续实验。使用Oligo7设计qPCR引物,以未处理、激素处理和病原菌处理的不同时间点的cDNA为模板,以EF1a为内参基因,在Rotor-Gene Q(QIAGEN)仪器上进行qPCR分析,实时荧光定量PCR体系为:TB GREENII酶10 μL,上下游引物各0.5 μL,木薯cDNA模板2 μL,ddH2O 7 μL,每个时间点设3个技术性重复。
-
以启动子区含有GCC-box这一顺式作用元件为条件,在木薯的全基因组数据中筛选符合条件的基因,共筛选到启动子区含有GCC-box这一顺式作用元件的基因204个,其基因详细信息见表1。
表 1 木薯ERF转录因子靶基因的注释
Table 1. Annotations of ERF transcription factor target gene in cassava
基因座位 Gene name 长度/bp Gene length 注释 Annotation Manes.01G064200 958 Osmotin-like protein OSM34 Manes.01G069100 3 335 Subtilase 4.12 Manes.02G024800 6 457 Transcription initiation factor TFIIE subunit beta Manes.02G189600 2 172 Putative WRKY transcription factor 33 Manes.03G039700 4 584 12-oxophytodienoate reductase 3 Manes.01G073700 3 652 Glycine cleavage T-protein family protein Manes.06G002000 6 512 Alpha/beta-hydrolase domain-containing protein Manes.09G063700 2 653 Phenylalanine ammonia-lyase 2 Manes.14G141600 9 106 Uncharacterized protein Manes.14G151300 386 Uncharacterized protein Manes.15G181900 4 511 Formate dehydrogenase Manes.18G073800 5 027 Histone-lysine N-methyltransferase ASHR1 Manes.01G079500 2 597 Uncharacterized protein Manes.01G091000 1 473 Hypothetical protein MANES_01G091000 Manes.01G105500 2 550 Alpha-1,4-galacturonosyltransferase Manes.01G121800 3 143 Uncharacterized protein Manes.01G132600 3 866 Uncharacterized protein Manes.01G133300 1 251 Uncharacterized protein Manes.01G155000 3 623 Protein kinase domain-containing protein Manes.01G156100 1 895 Pentatricopeptide repeat-containing protein Manes.01G164900 3 815 Laccase-4 Manes.01G171700 2 788 Scarecrow-like protein 34 Manes.01G191200 1 685 Protein TWIN LOV 1 Manes.01G194000 1 712 Myb-like transcription factor-like protein Manes.01G239000 10 955 Protein late elongated hypocotyl Manes.01G257700 623 RING/U-box domain-containing protein Manes.02G017600 1 568 Heavy-metal-associated domain-containing protein Manes.02G018800 1 163 Fiber Fb34 Manes.02G023700 101 Hypothetical protein MANES_02G023700 Manes.02G024900 11 644 Methyl- -binding domain-containing 13 isoform X1 Manes.02G036700 4 426 Adenine phosphoribosyl transferase 4 Manes.02G102800 1 121 GDP-mannose 4,6 dehydratase 2 Manes.02G108400 2 642 S-phase kinase-associated protein 1 Manes.02G121400 359 Josephin Manes.02G153300 3 946 Uncharacterized protein Manes.02G153900 2 548 Uncharacterized protein Manes.02G154000 1 220 Mitochondrial transcription termination factor family protein Manes.02G184300 760 Uncharacterized protein Manes.02G188500 635 Uncharacterized protein Manes.02G198700 128 Hypothetical protein MANES_02G198700 Manes.02G205000 146 Hypothetical protein MANES_02G205000 Manes.02G226700 2 598 U11/U12 small nuclear ribonucleoprotein Manes.03G012100 754 Uncharacterized protein Manes.03G032700 3 580 Methyltransferase Manes.03G036900 2 926 Protein phosphatase 2C 16 Manes.03G050700 11 341 AGC (cAMP-dependent, cGMP-dependent and protein kinase C)
kinase family proteinManes.03G051800 1 655 Mitochondrial substrate carrier family protein Manes.03G091000 1 460 Gibberellin 2-beta-dioxygenase 2 Manes.03G164100 3 813 Malate dehydrogenase Manes.03G183700 581 Aspartyl protease-like protein Manes.03G210000 2 101 Uncharacterized protein Manes.04G022300 197 Hypothetical protein MANES_04G022300 Manes.04G025100 5 961 Hydrolase, alpha/beta fold family protein Manes.04G035100 5 201 Uncharacterized protein Manes.05G033700 7 936 Signal peptide peptidase-like 4 Manes.05G057700 4 849 Alpha-1,3-glucosyltransferase Manes.05G062200 590 Plant invertase/pectin methylesterase inhibitor Domain-containing protein Manes.05G071300 636 Hypothetical protein MANES_05G071300 Manes.05G097100 10 203 Alpha-amylase-like 3 Manes.05G103400 2 351 Hypothetical protein MANES_05G103400, partial Manes.05G111500 2 499 Receptor-like protein kinase FERONIA Manes.05G119700 47 987 Uncharacterized protein Manes.05G159600 6 158 Uncharacterized protein Manes.05G174000 1 566 Sterol carrier protein 2 Manes.05G177200 2 505 VIER F-box protein 1 Manes.06G012700 2 821 Alternative oxidase 2 Manes.06G067300 3 592 Regulator of Vps4 activity in the MVB pathway protein Manes.06G078800 1 966 DNA-3-methyladenine glycosylase I Manes.06G129700 3 233 Glutamate dehydrogenase 2 Manes.06G140900 3 349 Protein kinase family protein Manes.06G164200 1 528 Chaperonin-like RbcX protein Manes.06G165700 839 Uncharacterized protein Manes.06G175500 2 430 Pentatricopeptide repeat-containing protein Manes.07G028100 6 262 Uncharacterized protein Manes.07G056000 9 058 Inositol-tetrakisphosphate 1-kinase 2 Manes.07G065200 1 549 F-box/kelch-repeat protein Manes.07G072000 7 499 Hypothetical protein MANES_07G072000 Manes.07G075200 2 214 Hypothetical protein MANES_07G075200 Manes.07G107500 1 805 Hypothetical protein MANES_07G107500 Manes.07G109000 1 895 Alpha/beta-hydrolase domain-containing protein Manes.07G129000 3 499 GDSL esterase/lipase CPRD49 Manes.07G129900 3 630 Haloacid dehalogenase-like hydrolase domain-containing protein Manes.07G131300 1 928 Putative protein phosphatase 2C 30 Manes.08G030000 1 009 Xanthoxin dehydrogenase Manes.08G039600 5 526 Translation initiation factor eIF-3 subunit 8 Manes.08G108800 2 055 Pectin lyase-like protein Manes.08G136100 662 Uncharacterized protein Manes.08G137700 947 Ethylene-responsive transcription factor 1B Manes.08G147800 1 983 NAC domain containing protein 47 Manes.08G148900 3 450 Uncharacterized protein Manes.08G162000 338 Hypothetical protein MANES_08G162000 Manes.09G008400 5 230 Ankyrin repeat and BTB/POZ domain-containing protein Manes.09G009700 622 Hypothetical protein MANES_09G009700 Manes.09G107500 1 883 PHD finger-like domain-containing protein 5A Manes.09G109400 7 454 Tetratricopeptide repeat (TPR)-containing protein Manes.09G146900 6 045 Heme oxygenase-like, multi-helical protein Manes.09G149000 1 173 Ethylene-responsive transcription factor 1B Manes.09G165800 1 122 Rossmann-fold NAD(P)-binding domain-containing protein Manes.09G185400 4 108 Pyruvate dehydrogenase E1 component subunit beta Manes.10G002300 10 867 F-box domain-containing protein Manes.10G004700 6 026 Potassium transporter 2 Manes.10G007300 5 373 Biotin/lipoyl attachment domain-containing protein Manes.10G041100 3 379 Floral homeotic protein APETALA 2 Manes.10G072100 1 751 60S ribosomal protein L8-3 Manes.10G094000 2 043 TTF-type zinc finger protein with HAT dimerisation domain Manes.10G097300 10 362 Dynamin-related protein 3A Manes.10G111800 7 592 Zinc finger CCCH domain-containing protein 5 Manes.10G119500 4 800 Ethylene-responsive transcription factor RAP2-7 Manes.10G123900 368 Uncharacterized protein Manes.11G005600 1 016 NADPH:quinone oxidoreductase Manes.11G009200 1 991 Xyloglucan fucosyltransferase Manes.11G014100 1 440 GDSL esterase/lipase 1 Manes.11G019800 5 849 E3 ubiquitin ligase-like protein Manes.11G022100 5 549 Phospholipid:diacylglycerol acyltransferase 1 Manes.11G040800 1 832 Protein kinase family protein Manes.11G058500 5 067 Prefoldin 6 Manes.11G067200 1 153 Lamin-like protein Manes.11G070400 2 939 Wall-associated receptor kinase-like 22 Manes.11G071000 1 085 T-complex protein 1 subunit epsilon Manes.11G105000 3 083 Elongation factor EF-2 Manes.11G141100 2 601 GTP binding protein Manes.12G007000 416 Glutaredoxin-C8 Manes.12G009300 2 317 Uncharacterized protein Manes.12G024200 4 084 Ribose-phosphate pyrophosphokinase 3 Manes.12G043700 3 925 26S proteasome regulatory subunit 4-A Manes.12G059000 1 090 ILI1 binding bHLH 1 protein Manes.12G066300 1 289 Nuclear transcription factor Y subunit B-3 Manes.12G095300 953 Dof zinc finger protein DOF3.4 Manes.12G096300 1 524 Dof zinc finger protein DOF1.1 Manes.12G110100 2 614 Acyl-CoA N-acyltransferases-like protein Manes.12G111300 3 013 Phototropic-responsive NPH3-like protein Manes.12G114300 1 921 Cytochrome P450, family 81, subfamily D, polypeptide 5 Manes.12G138000 215 Hypothetical protein MANES_12G138000 Manes.13G007200 416 Glutaredoxin-C7 Manes.13G007300 1 265 Pollen Ole e 1 allergen and extensin family protein Manes.13G029700 4 707 Protein kinase family protein Manes.13G117100 2 242 Cytochrome P450, family 81, subfamily D, polypeptide 8 Manes.13G150000 7 593 STRUBBELIG-receptor family 3 Manes.14G001200 3 318 Hypothetical protein MANES_14G001200 Manes.14G006900 4 475 Mitochondrial transcription termination factor family protein Manes.14G024500 5 322 SKP1-like protein 21 Manes.14G036100 4 095 LUC7 N_terminus domain-containing protein Manes.14G071200 621 Hypothetical protein MANES_14G071200 Manes.14G112300 3 289 Uncharacterized protein Manes.14G125900 877 Transcription factor bHLH52 Manes.14G134200 1 024 Protein LURP-one-related 12 Manes.15G018300 2 852 Hypothetical protein MANES_15G018300 Manes.15G023900 1 418 Aspartyl protease-like protein Manes.15G050500 1 913 9-cis-epoxycarotenoid dioxygenase NCED6 Manes.15G053800 5 533 High mobility group-box and ARID domain-binding domain-containing protein Manes.15G070500 479 myb domain protein 101 Manes.15G090000 997 Homeobox-leucine zipper protein ATHB-52 Manes.15G103900 1 499 Peroxidase 59 Manes.15G109900 269 Hypothetical protein MANES_15G109900 Manes.15G134200 5 566 UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase Manes.15G150600 884 Uncharacterized protein Manes.15G152100 5 554 SGF29 tudor-like domain-containing protein Manes.15G166600 3 570 Putative cysteine-rich receptor-like protein kinase 43 Manes.15G179800 7 127 Alpha-mannosidase II Manes.16G007600 650 Uncharacterized protein Manes.16G020700 1 432 F-box protein Manes.16G054300 4 718 Galacturonosyltransferase 6 Manes.16G056200 1 763 TTF-type zinc finger protein with HAT dimerization domain Manes.16G058300 2 179 TTF-type zinc finger protein with HAT dimerization domain Manes.16G066800 558 Uncharacterized protein Manes.16G077300 2 074 Uncharacterized protein Manes.16G083800 1 764 Mitochondrial substrate carrier family protein Manes.16G084600 2 554 Thermospermine synthase ACAULIS5 Manes.16G087300 782 Ethylene-responsive transcription factor ERF016 Manes.16G109800 4 580 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase-like protein Manes.16G116900 554 Hypothetical protein MANES_16G116900 Manes.17G000400 23 515 Nuclear RNA polymerase C1 Manes.17G024800 248 Hypothetical protein MANES_17G024800 Manes.17G024900 398 Hypothetical protein MANES_17G024900, partial Manes.17G030400 2 314 Phototropic-responsive NPH3 family protein Manes.17G048000 518 Hypothetical protein MANES_17G048000 Manes.17G048400 746 Ethylene-responsive transcription factor 11 Manes.17G048500 6 168 MAC/Perforin domain-containing protein Manes.17G056700 5 022 Uncharacterized protein Manes.17G073100 4 620 Endoplasmic oxidoreductin-2 Manes.17G112300 996 Uncharacterized protein Manes.17G112400 20 885 Hypothetical protein MANES_17G112400 Manes.18G001100 6 966 Pyrophosphate--fructose-6-phosphate 1-phosphotransferase Manes.18G011500 2 613 Adenine nucleotide alpha hydrolases-domain containing protein kinase Manes.18G011600 321 Hypothetical protein MANES_18G011600 Manes.18G011700 11 468 Vacuolar sorting protein 35 Manes.18G047000 2 741 Transcription factor lim1 Manes.18G052700 2 457 DnaJ/Hsp40 cysteine-rich domain-containing protein Manes.18G058800 10 440 Kinesin family member 4/7/21/27 Manes.18G098700 2 548 Putative WRKY transcription factor 33 Manes.S014400 2 771 Disease resistance-like protein/LRR domain-containing protein Manes.S014600 2 975 Disease resistance-like protein/LRR domain-containing protein Manes.S014800 14 231 Disease resistance-like protein/LRR domain-containing protein Manes.S020700 1 380 Myb domain protein 93 Manes.S027500 1 737 F-box/kelch-repeat protein Manes.S030800 4 132 RAB GTPase homolog A5A Manes.S054400 1 964 Glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein Manes.S054500 4 127 Polygalacturonase-like protein Manes.S054600 1 461 Polygalacturonase-like protein Manes.S054700 1 461 Polygalacturonase-like protein Manes.S055000 1 461 Polygalacturonase-like protein Manes.S055100 1 461 polygalacturonase-like protein Manes.S096700 3 378 Uncharacterized protein Manes.S109400 929 Polygalacturonase-like protein -
对204个木薯ERF转录因子靶基因的染色体位置分布进行分析,结果见图1。从图1可知,有15个基因定位于未知的染色体上,剩余189个基因在木薯的18条染色体上均有分布。其中,木薯第2条染色体上有18个基因,是分布最多的染色体;第4条染色体上只有3个基因,是分布最少的染色体。位于染色体上端的基因最多,占43%,第4条和第18条染色体上,基因均分布在染色体的上端,中部和下端没有分布;位于染色体下端的基因占35%;位于染色体中部的基因仅占22%。
-
木薯ERF转录因子靶基因的注释中,Manes.02G189600与拟南芥中的AtWRKY33基因同源,而AtWRKY33是植物病原菌互作途径中重要转录因子;Manes.03G039700是OPR3,JA信号通路中的同工酶;Manes.09G063700是苯丙烷类代谢途径的关键酶,也是合成木质素的关键酶;Manes.15G181900是甲酸氢解酶;而其他大部分基因编码的蛋白是未知蛋白。
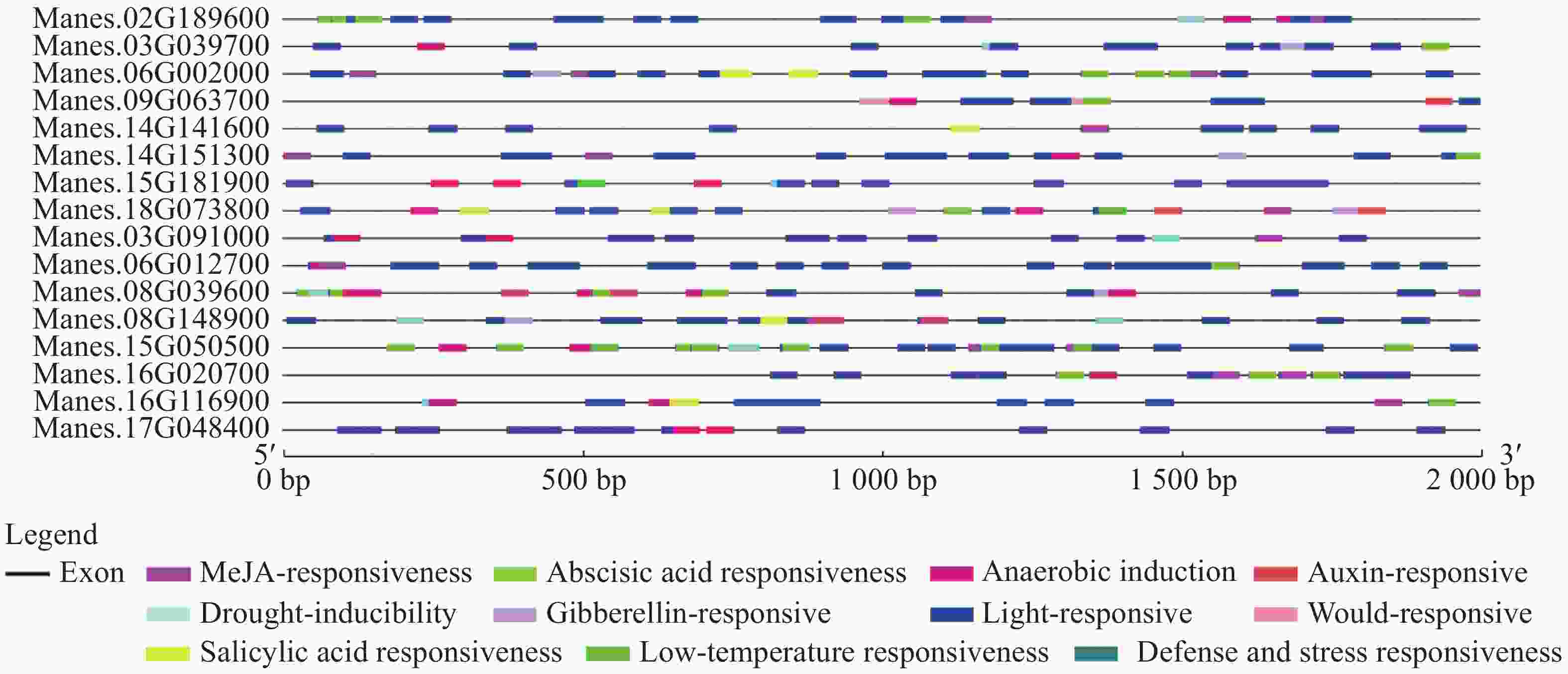
选取上述4个基因,再随机挑选12个基因,通过分析这16个基因的启动子顺式作用元件,发现其具有典型的启动子元件TATA-box和CAAT-box,说明具有启动子活性(图2)。从图2进一步分析发现,有大量的光响应元件,例如ACE,G-box,Box 4,MRE等;还存在防御和胁迫响应元件(TC-rich repeats)、低温响应元件(LTR)、干旱响应元件(MBS)、脱落酸响应元件(ABRE)、茉莉酸甲酯响应元件(TGACG-motif,CGTCA-motif)等。多种顺式作用元件的存在说明这些基因可能受到复杂的调控,在木薯遭遇胁迫时发挥一定的作用。
-
在NCBI网站SRA数据库中,下载来源于用Xam弱致病菌株ORST4和强致病菌株ORST4+TALE1侵染苗龄8周的木薯(品种为MCOL1522)组培苗实验的12个关于病原菌侵染木薯的转录组数据[17],详见表2。
表 2 病原菌侵染木薯的转录组数据来源
Table 2. Transcriptome data sources of cassava infected by pathogenic bacteria
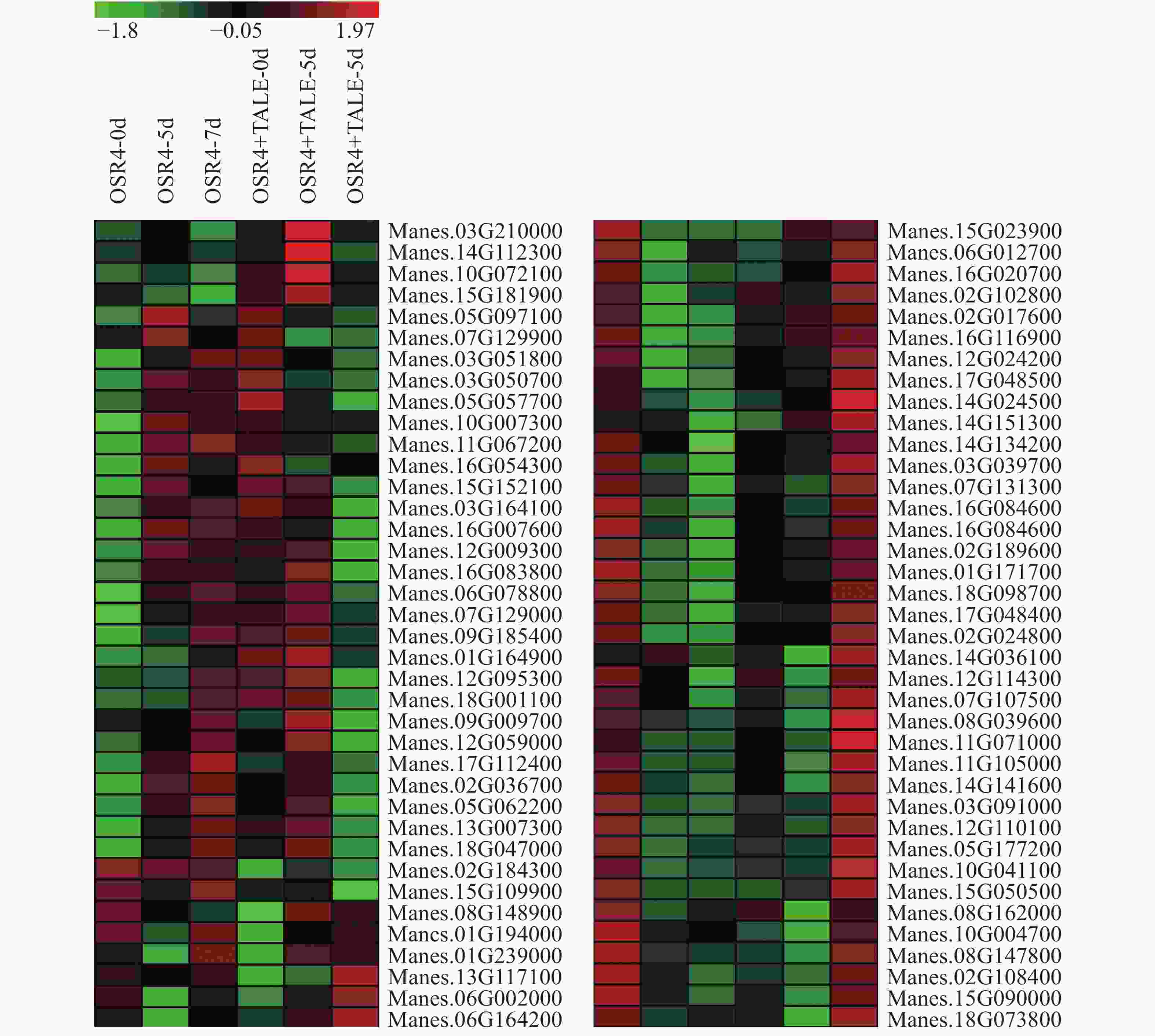
运行ID Run ID 样品名称 Sampling SRR1050891 ORST4, collected at 0 d SRR1050892 ORST4, collected at 5 d SRR1050893 ORST4, collected at 7 d SRR1050894 ORST4+TALE1Xam, collected at 0 d SRR1050895 ORST4+TALE1Xam, collected at 5 d SRR1050896 ORST4+TALE1Xam, collected at 7 d SRR1050897 ORST4, collected at 0 d SRR1050898 ORST4, collected at 5 d SRR1050899 ORST4, collected at 7 d SRR1050900 ORST4+TALE1Xam, collected at 0 d SRR1050901 ORST4+TALE1Xam, collected at 5 d SRR1050902 ORST4+TALE1Xam, collected at 7 d 计算木薯ERF转录因子靶基因在这些转录组中的FPKM值,表达量低于1的基因剔除,筛选出76个基因用Heatmap构建基因表达热图(图3)。从图3可知,有26个基因能够被强致病菌株诱导表达,被弱致病菌株抑制表达,其中,Manes.03G039700,Manes.06G002000,Manes.14G141600,Manes.15G181900,Manes.03G091000,Manes.06G012700,Manes.08G039600,Manes.08G148900,Manes.15G050500,Manes.16G020700,Manes.16G116900,Manes.17G048400这12个基因的表达差异最为显著,尤其是Manes.03G091000,在病原菌侵染前后基因的表达差异超过了75倍。
-
选取Manes.02G189600,Manes.03G039700,Manes.06G002000,Manes.09G063700,Manes.14G141600,Manes.15G181900等6个基因,利用qRT-PCR作进一步验证。
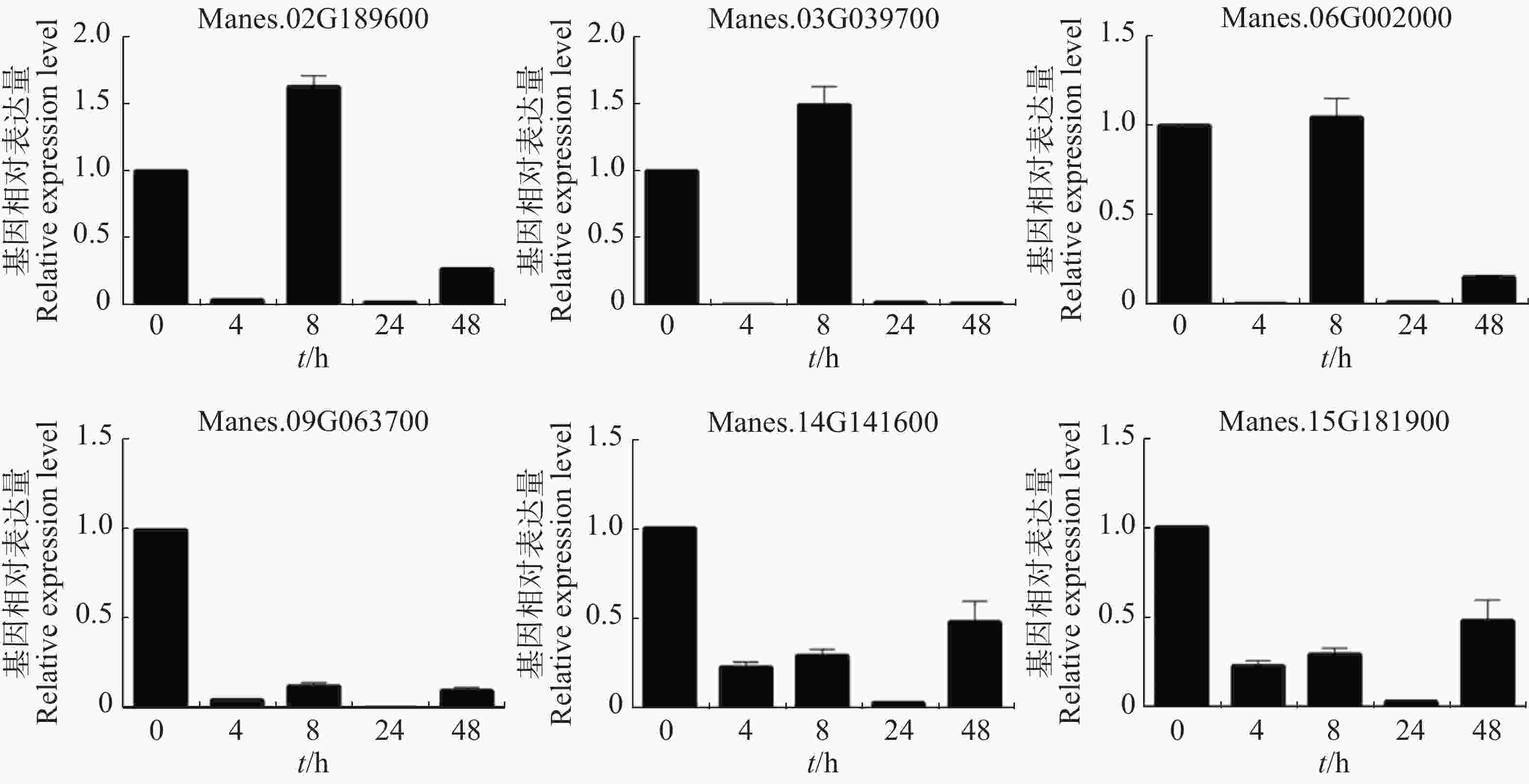
用ACC处理木薯幼苗,从图4可知,Manes.02G189600,Manes.03G039700和Manes.15G181900基因在处理0.5 h后表达量均持续上调。其中,Manes.03G039700基因表达量上调更显著;Manes.02G189600基因在处理0.5 h后表达量最高,之后上调缓慢;Manes.15G181900基因在处理6 h后表达量最高,上调较显著。Manes.06G002000基因在处理后表达量下调,1 h表达量最低,之后表达量(相比1 h)上调,6 h的表达量与0 h几乎一致。Manes.09G063700 和Manes.14G141600基因在处理1 h后表达量最低,而2 h的表达量回升达到最高,之后Manes.09G063700基因表达量与0 h几乎一致,Manes.14G141600基因上调缓慢。本实验结果表明,这6个基因可以瞬时响应ACC处理,可能在木薯的乙烯通路中发挥作用。
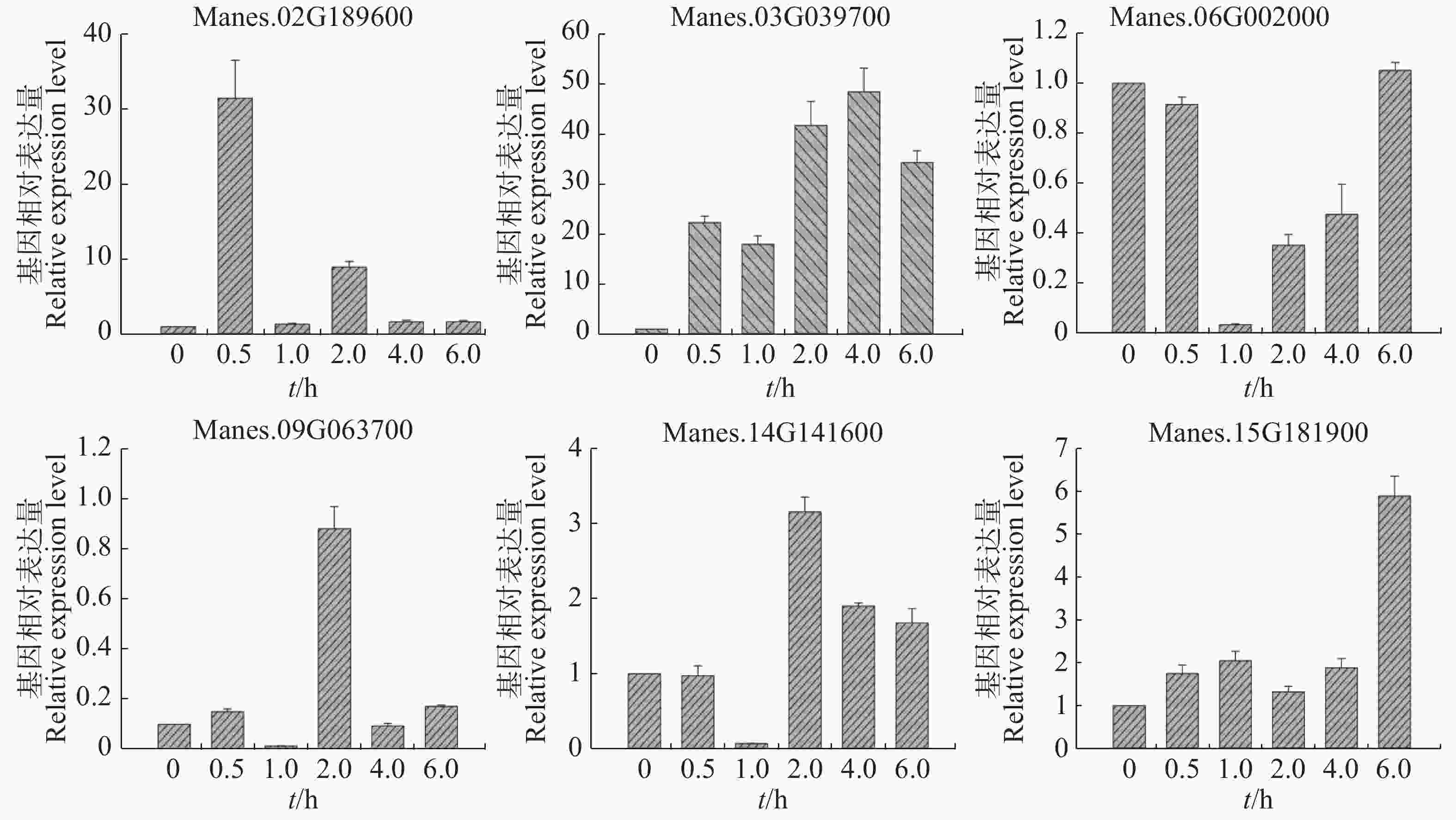
图 4 木薯ERF转录因子靶基因在ACC处理下的表达模式
Figure 4. Expression pattern of ERF transcription factor target genes in cassava under ACC treatment
从图5可知,在病原菌侵染木薯幼苗4 h后(相比对照),Manes.02G189600,Manes.03G039700,Manes.06G002000,Manes.09G063700,Manes.14G141600,Manes.15G181900基因的表达量均显著下调;Manes.02G189600,Manes.03G039700和Manes.06G002000在病原菌侵染8 h(相比对照)的表达量上调;24 h后(相比对照),Manes.02G189600,Manes.03G039700,Manes.06G002000,Manes.09G063700,Manes.14G141600,Manes.15G181900这6个基因的表达量继续下调。本实验结果表明,这些基因可以瞬时响应病原菌对木薯的侵染,可能参与了木薯的抗病途径。
-
在对各种外界胁迫的适应过程中,植物体内发生了一系列的信号传递,并激发其防御体系,使植物产生防卫反应,提高其自身抵抗生物和非生物胁迫的能力。这些防卫反应涉及大量相关基因在转录水平上的调整,其中,各种转录因子与其目标基因启动子上顺式作用元件的识别和结合起关键作用[18-19]。GCC-box通常存在于植物PR基因的启动子中,如在拟南芥的PDF2.1、烟草的PR3等基因的启动子中均含有GCC-box,在一些非生物胁迫应答基因的启动子中也含有GCC-box[11, 20-21]。番茄ERF2能分别结合乙烯合成相关基因ACS3和ACO3启动子中的GCC盒和DRE顺式作用元件,调控乙烯的合成[22]。在过表达ERF104的拟南芥转基因植株的基因表达谱分析中,发现534个表达水平上调3倍以上的基因,包括PDF1.2,PR5,MKK4,RBOHD,ERF4,WRKY33和TGA1.3等一些已知的防卫基因或抗病信号调控基因,其中,有2个PDF1.2基因的表达水平升高近1 000倍,而表达水平上调10倍以上的基因启动子中含有GCC-box[23]。GCC-box是ERF转录因子的主要顺式作用元件,一些ERF转录因子能通过结合启动子中GCC-box来调控靶标基因的表达。GCC-box存在于大量的PR基因启动子中,ERF转录因子可以通过结合GCC-box直接调节PR基因的表达,如拟南芥ERF1和ORA59能直接与PR基因PDF1.2启动子中的GCC顺式作用元件结合,并激活其表达[24]。
本研究从木薯全基因组中筛选出了204个启动子区含有GCC-box顺式作用元件的基因,它们在木薯的18条染色体上均有分布。通过分析这些基因在病原菌侵染下的表达模式,从中选取表达差异比较显著的基因,并对它们的启动子区顺式作用元件进行预测分析,发现这些基因的启动子区都含有多种胁迫响应元件,多种顺式作用元件的存在说明这些基因可能受到复杂的调控,在木薯遭遇胁迫时发挥一定的作用。本研究利用qPCR验证的结果进一步表明,这些基因可以响应病原菌对木薯的侵染,初步推断这些基因可能参与了木薯的抗病途径。本实验结果为木薯抗病机制及抗病育种研究奠定了良好的基础。
Characterization and Expression Analysis of the Target Genes Regulated by ERF in Cassava
-
摘要: 为解析乙烯响应因子(ERF, Ethylene Response Factor)在木薯(Manihot esculenta)生长发育过程中的调控通路,本研究从木薯全基因组数据库中,筛选出启动子区含有GCC-box顺式作用元件的基因204个,利用生物信息学手段对这些基因进行染色体位置分布分析、病原菌侵染下表达模式分析以及启动子结构预测,部分候选基因的qRT-PCR结果表明:Manes.02G189600,Manes.03G039700,Manes.06G002000,Manes.09G063700,Manes.14G141600,Manes.15G181900可能参与调控植物的抗病途径,同时在乙烯介导的信号传递途径中也起重要作用。Abstract: The defense response of plants to stress is largely regulated by the level of gene transcription, and the recognition and combination of the transcription factor and cis-acting elements on the target gene promoter plays a key regulatory role. ERF is a type of transcription factor that plays an important role in the growth and development of plants. There is a conserved GCC-box in the promoter region of downstream target genes regulated by it. In order to analyze the regulatory pathway of ERF in the growth and development of cassava, 204 genes containing GCC-box cis-acting elements in the promoter region were screened. Chromosomal position distribution, analysis of expression patterns under pathogen infection, and promoter structure prediction of these genes were analyzed using bioinformatics methods. The qRT-PCR results showed that Manes.02G189600, Manes.03G039700, Manes.06G002000, Manes.09G063700, Manes.14G141600, Manes.15G181900 might be involved in regulating plant disease resistance pathways, and play an important way in ethylene-mediated signal transduction pathways.
-
Key words:
- cassava /
- GCC-box /
- ERF transcription factor /
- biological stress
-
表 1 木薯ERF转录因子靶基因的注释
Table 1 Annotations of ERF transcription factor target gene in cassava
基因座位 Gene name 长度/bp Gene length 注释 Annotation Manes.01G064200 958 Osmotin-like protein OSM34 Manes.01G069100 3 335 Subtilase 4.12 Manes.02G024800 6 457 Transcription initiation factor TFIIE subunit beta Manes.02G189600 2 172 Putative WRKY transcription factor 33 Manes.03G039700 4 584 12-oxophytodienoate reductase 3 Manes.01G073700 3 652 Glycine cleavage T-protein family protein Manes.06G002000 6 512 Alpha/beta-hydrolase domain-containing protein Manes.09G063700 2 653 Phenylalanine ammonia-lyase 2 Manes.14G141600 9 106 Uncharacterized protein Manes.14G151300 386 Uncharacterized protein Manes.15G181900 4 511 Formate dehydrogenase Manes.18G073800 5 027 Histone-lysine N-methyltransferase ASHR1 Manes.01G079500 2 597 Uncharacterized protein Manes.01G091000 1 473 Hypothetical protein MANES_01G091000 Manes.01G105500 2 550 Alpha-1,4-galacturonosyltransferase Manes.01G121800 3 143 Uncharacterized protein Manes.01G132600 3 866 Uncharacterized protein Manes.01G133300 1 251 Uncharacterized protein Manes.01G155000 3 623 Protein kinase domain-containing protein Manes.01G156100 1 895 Pentatricopeptide repeat-containing protein Manes.01G164900 3 815 Laccase-4 Manes.01G171700 2 788 Scarecrow-like protein 34 Manes.01G191200 1 685 Protein TWIN LOV 1 Manes.01G194000 1 712 Myb-like transcription factor-like protein Manes.01G239000 10 955 Protein late elongated hypocotyl Manes.01G257700 623 RING/U-box domain-containing protein Manes.02G017600 1 568 Heavy-metal-associated domain-containing protein Manes.02G018800 1 163 Fiber Fb34 Manes.02G023700 101 Hypothetical protein MANES_02G023700 Manes.02G024900 11 644 Methyl- -binding domain-containing 13 isoform X1 Manes.02G036700 4 426 Adenine phosphoribosyl transferase 4 Manes.02G102800 1 121 GDP-mannose 4,6 dehydratase 2 Manes.02G108400 2 642 S-phase kinase-associated protein 1 Manes.02G121400 359 Josephin Manes.02G153300 3 946 Uncharacterized protein Manes.02G153900 2 548 Uncharacterized protein Manes.02G154000 1 220 Mitochondrial transcription termination factor family protein Manes.02G184300 760 Uncharacterized protein Manes.02G188500 635 Uncharacterized protein Manes.02G198700 128 Hypothetical protein MANES_02G198700 Manes.02G205000 146 Hypothetical protein MANES_02G205000 Manes.02G226700 2 598 U11/U12 small nuclear ribonucleoprotein Manes.03G012100 754 Uncharacterized protein Manes.03G032700 3 580 Methyltransferase Manes.03G036900 2 926 Protein phosphatase 2C 16 Manes.03G050700 11 341 AGC (cAMP-dependent, cGMP-dependent and protein kinase C)
kinase family proteinManes.03G051800 1 655 Mitochondrial substrate carrier family protein Manes.03G091000 1 460 Gibberellin 2-beta-dioxygenase 2 Manes.03G164100 3 813 Malate dehydrogenase Manes.03G183700 581 Aspartyl protease-like protein Manes.03G210000 2 101 Uncharacterized protein Manes.04G022300 197 Hypothetical protein MANES_04G022300 Manes.04G025100 5 961 Hydrolase, alpha/beta fold family protein Manes.04G035100 5 201 Uncharacterized protein Manes.05G033700 7 936 Signal peptide peptidase-like 4 Manes.05G057700 4 849 Alpha-1,3-glucosyltransferase Manes.05G062200 590 Plant invertase/pectin methylesterase inhibitor Domain-containing protein Manes.05G071300 636 Hypothetical protein MANES_05G071300 Manes.05G097100 10 203 Alpha-amylase-like 3 Manes.05G103400 2 351 Hypothetical protein MANES_05G103400, partial Manes.05G111500 2 499 Receptor-like protein kinase FERONIA Manes.05G119700 47 987 Uncharacterized protein Manes.05G159600 6 158 Uncharacterized protein Manes.05G174000 1 566 Sterol carrier protein 2 Manes.05G177200 2 505 VIER F-box protein 1 Manes.06G012700 2 821 Alternative oxidase 2 Manes.06G067300 3 592 Regulator of Vps4 activity in the MVB pathway protein Manes.06G078800 1 966 DNA-3-methyladenine glycosylase I Manes.06G129700 3 233 Glutamate dehydrogenase 2 Manes.06G140900 3 349 Protein kinase family protein Manes.06G164200 1 528 Chaperonin-like RbcX protein Manes.06G165700 839 Uncharacterized protein Manes.06G175500 2 430 Pentatricopeptide repeat-containing protein Manes.07G028100 6 262 Uncharacterized protein Manes.07G056000 9 058 Inositol-tetrakisphosphate 1-kinase 2 Manes.07G065200 1 549 F-box/kelch-repeat protein Manes.07G072000 7 499 Hypothetical protein MANES_07G072000 Manes.07G075200 2 214 Hypothetical protein MANES_07G075200 Manes.07G107500 1 805 Hypothetical protein MANES_07G107500 Manes.07G109000 1 895 Alpha/beta-hydrolase domain-containing protein Manes.07G129000 3 499 GDSL esterase/lipase CPRD49 Manes.07G129900 3 630 Haloacid dehalogenase-like hydrolase domain-containing protein Manes.07G131300 1 928 Putative protein phosphatase 2C 30 Manes.08G030000 1 009 Xanthoxin dehydrogenase Manes.08G039600 5 526 Translation initiation factor eIF-3 subunit 8 Manes.08G108800 2 055 Pectin lyase-like protein Manes.08G136100 662 Uncharacterized protein Manes.08G137700 947 Ethylene-responsive transcription factor 1B Manes.08G147800 1 983 NAC domain containing protein 47 Manes.08G148900 3 450 Uncharacterized protein Manes.08G162000 338 Hypothetical protein MANES_08G162000 Manes.09G008400 5 230 Ankyrin repeat and BTB/POZ domain-containing protein Manes.09G009700 622 Hypothetical protein MANES_09G009700 Manes.09G107500 1 883 PHD finger-like domain-containing protein 5A Manes.09G109400 7 454 Tetratricopeptide repeat (TPR)-containing protein Manes.09G146900 6 045 Heme oxygenase-like, multi-helical protein Manes.09G149000 1 173 Ethylene-responsive transcription factor 1B Manes.09G165800 1 122 Rossmann-fold NAD(P)-binding domain-containing protein Manes.09G185400 4 108 Pyruvate dehydrogenase E1 component subunit beta Manes.10G002300 10 867 F-box domain-containing protein Manes.10G004700 6 026 Potassium transporter 2 Manes.10G007300 5 373 Biotin/lipoyl attachment domain-containing protein Manes.10G041100 3 379 Floral homeotic protein APETALA 2 Manes.10G072100 1 751 60S ribosomal protein L8-3 Manes.10G094000 2 043 TTF-type zinc finger protein with HAT dimerisation domain Manes.10G097300 10 362 Dynamin-related protein 3A Manes.10G111800 7 592 Zinc finger CCCH domain-containing protein 5 Manes.10G119500 4 800 Ethylene-responsive transcription factor RAP2-7 Manes.10G123900 368 Uncharacterized protein Manes.11G005600 1 016 NADPH:quinone oxidoreductase Manes.11G009200 1 991 Xyloglucan fucosyltransferase Manes.11G014100 1 440 GDSL esterase/lipase 1 Manes.11G019800 5 849 E3 ubiquitin ligase-like protein Manes.11G022100 5 549 Phospholipid:diacylglycerol acyltransferase 1 Manes.11G040800 1 832 Protein kinase family protein Manes.11G058500 5 067 Prefoldin 6 Manes.11G067200 1 153 Lamin-like protein Manes.11G070400 2 939 Wall-associated receptor kinase-like 22 Manes.11G071000 1 085 T-complex protein 1 subunit epsilon Manes.11G105000 3 083 Elongation factor EF-2 Manes.11G141100 2 601 GTP binding protein Manes.12G007000 416 Glutaredoxin-C8 Manes.12G009300 2 317 Uncharacterized protein Manes.12G024200 4 084 Ribose-phosphate pyrophosphokinase 3 Manes.12G043700 3 925 26S proteasome regulatory subunit 4-A Manes.12G059000 1 090 ILI1 binding bHLH 1 protein Manes.12G066300 1 289 Nuclear transcription factor Y subunit B-3 Manes.12G095300 953 Dof zinc finger protein DOF3.4 Manes.12G096300 1 524 Dof zinc finger protein DOF1.1 Manes.12G110100 2 614 Acyl-CoA N-acyltransferases-like protein Manes.12G111300 3 013 Phototropic-responsive NPH3-like protein Manes.12G114300 1 921 Cytochrome P450, family 81, subfamily D, polypeptide 5 Manes.12G138000 215 Hypothetical protein MANES_12G138000 Manes.13G007200 416 Glutaredoxin-C7 Manes.13G007300 1 265 Pollen Ole e 1 allergen and extensin family protein Manes.13G029700 4 707 Protein kinase family protein Manes.13G117100 2 242 Cytochrome P450, family 81, subfamily D, polypeptide 8 Manes.13G150000 7 593 STRUBBELIG-receptor family 3 Manes.14G001200 3 318 Hypothetical protein MANES_14G001200 Manes.14G006900 4 475 Mitochondrial transcription termination factor family protein Manes.14G024500 5 322 SKP1-like protein 21 Manes.14G036100 4 095 LUC7 N_terminus domain-containing protein Manes.14G071200 621 Hypothetical protein MANES_14G071200 Manes.14G112300 3 289 Uncharacterized protein Manes.14G125900 877 Transcription factor bHLH52 Manes.14G134200 1 024 Protein LURP-one-related 12 Manes.15G018300 2 852 Hypothetical protein MANES_15G018300 Manes.15G023900 1 418 Aspartyl protease-like protein Manes.15G050500 1 913 9-cis-epoxycarotenoid dioxygenase NCED6 Manes.15G053800 5 533 High mobility group-box and ARID domain-binding domain-containing protein Manes.15G070500 479 myb domain protein 101 Manes.15G090000 997 Homeobox-leucine zipper protein ATHB-52 Manes.15G103900 1 499 Peroxidase 59 Manes.15G109900 269 Hypothetical protein MANES_15G109900 Manes.15G134200 5 566 UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase Manes.15G150600 884 Uncharacterized protein Manes.15G152100 5 554 SGF29 tudor-like domain-containing protein Manes.15G166600 3 570 Putative cysteine-rich receptor-like protein kinase 43 Manes.15G179800 7 127 Alpha-mannosidase II Manes.16G007600 650 Uncharacterized protein Manes.16G020700 1 432 F-box protein Manes.16G054300 4 718 Galacturonosyltransferase 6 Manes.16G056200 1 763 TTF-type zinc finger protein with HAT dimerization domain Manes.16G058300 2 179 TTF-type zinc finger protein with HAT dimerization domain Manes.16G066800 558 Uncharacterized protein Manes.16G077300 2 074 Uncharacterized protein Manes.16G083800 1 764 Mitochondrial substrate carrier family protein Manes.16G084600 2 554 Thermospermine synthase ACAULIS5 Manes.16G087300 782 Ethylene-responsive transcription factor ERF016 Manes.16G109800 4 580 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase-like protein Manes.16G116900 554 Hypothetical protein MANES_16G116900 Manes.17G000400 23 515 Nuclear RNA polymerase C1 Manes.17G024800 248 Hypothetical protein MANES_17G024800 Manes.17G024900 398 Hypothetical protein MANES_17G024900, partial Manes.17G030400 2 314 Phototropic-responsive NPH3 family protein Manes.17G048000 518 Hypothetical protein MANES_17G048000 Manes.17G048400 746 Ethylene-responsive transcription factor 11 Manes.17G048500 6 168 MAC/Perforin domain-containing protein Manes.17G056700 5 022 Uncharacterized protein Manes.17G073100 4 620 Endoplasmic oxidoreductin-2 Manes.17G112300 996 Uncharacterized protein Manes.17G112400 20 885 Hypothetical protein MANES_17G112400 Manes.18G001100 6 966 Pyrophosphate--fructose-6-phosphate 1-phosphotransferase Manes.18G011500 2 613 Adenine nucleotide alpha hydrolases-domain containing protein kinase Manes.18G011600 321 Hypothetical protein MANES_18G011600 Manes.18G011700 11 468 Vacuolar sorting protein 35 Manes.18G047000 2 741 Transcription factor lim1 Manes.18G052700 2 457 DnaJ/Hsp40 cysteine-rich domain-containing protein Manes.18G058800 10 440 Kinesin family member 4/7/21/27 Manes.18G098700 2 548 Putative WRKY transcription factor 33 Manes.S014400 2 771 Disease resistance-like protein/LRR domain-containing protein Manes.S014600 2 975 Disease resistance-like protein/LRR domain-containing protein Manes.S014800 14 231 Disease resistance-like protein/LRR domain-containing protein Manes.S020700 1 380 Myb domain protein 93 Manes.S027500 1 737 F-box/kelch-repeat protein Manes.S030800 4 132 RAB GTPase homolog A5A Manes.S054400 1 964 Glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein Manes.S054500 4 127 Polygalacturonase-like protein Manes.S054600 1 461 Polygalacturonase-like protein Manes.S054700 1 461 Polygalacturonase-like protein Manes.S055000 1 461 Polygalacturonase-like protein Manes.S055100 1 461 polygalacturonase-like protein Manes.S096700 3 378 Uncharacterized protein Manes.S109400 929 Polygalacturonase-like protein 表 2 病原菌侵染木薯的转录组数据来源
Table 2 Transcriptome data sources of cassava infected by pathogenic bacteria
运行ID Run ID 样品名称 Sampling SRR1050891 ORST4, collected at 0 d SRR1050892 ORST4, collected at 5 d SRR1050893 ORST4, collected at 7 d SRR1050894 ORST4+TALE1Xam, collected at 0 d SRR1050895 ORST4+TALE1Xam, collected at 5 d SRR1050896 ORST4+TALE1Xam, collected at 7 d SRR1050897 ORST4, collected at 0 d SRR1050898 ORST4, collected at 5 d SRR1050899 ORST4, collected at 7 d SRR1050900 ORST4+TALE1Xam, collected at 0 d SRR1050901 ORST4+TALE1Xam, collected at 5 d SRR1050902 ORST4+TALE1Xam, collected at 7 d -
[1] HERNANDEZ-GARCIA C M, FINER J J. Identification and validation of promoters and cis-acting regulatory elements [J]. Plant Science: an international journal of experimental plant biology, 2014, 217: 109 − 119. [2] MITSUDA N, OHME-TAKAGI M. Functional analysis of transcription factors in Arabidopsis [J]. Plant and Cell Physiology, 2009, 50(7): 1232 − 1248. doi: 10.1093/pcp/pcp075 [3] CHEN W, PROVART N J, JA NE, et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses [J]. The Plant cell, 2002, 14(3): 559 − 574. doi: 10.1105/tpc.010410 [4] CHEONG Y H, CHANG H -S, GUPTA R, et al. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis [J]. Plant physiology, 2002, 129(2): 661 − 677. doi: 10.1104/pp.002857 [5] SCHENK P M, KAZAN K, WILSON I, et al. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis [J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(21): 11655 − 11660. doi: 10.1073/pnas.97.21.11655 [6] FRANCO-ZORRILLA J M, LOPEZ-VIDRIERO I, CARRASCO J L, et al. DNA-binding specificities of plant transcription factors and their potential to define target genes [J]. Proceedings of the National Academy of Sciences, 2014, 111(6): 2367 − 2372. doi: 10.1073/pnas.1316278111 [7] EINI O, YANG N, PYVOVARENKO T, et al. Complex regulation by Apetala2 domain-containing transcription factors revealed through analysis of the stress-responsive TdCor410b promoter from durum wheat [J]. PLoS One, 2013, 8(3): e58713. doi: 10.1371/journal.pone.0058713 [8] OHME-TAKAGI M and SHINSHI H. Ethylene- inducible DNA binding proteins that interact with an ethylene- responsive element [J]. The Plant cell, 1995, 7(2): 173 − 182. [9] DONG W, AI X, XU F, et al. Isolation and characterization of a bread wheat salinity responsive ERF transcription factor [J]. Gene, 2012, 511(1): 38 − 45. doi: 10.1016/j.gene.2012.09.039 [10] FUJIMOTO S Y, OHTA M, USUI A, et al. Arabidopsis ethylene- responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression [J]. The Plant cell, 2000, 12(3): 393 − 404. [11] BUTTNER M, SINGH K B. Arabidopsis thaliana ethylene- responsive element binding protein (AtEBP), an ethylene- inducible, GCC box DNA- binding protein interacts with an ocs element binding protein [J]. Proceedings of the National Academy of Sciences of the United States of America, 1997, 94(11): 5961 − 5966. doi: 10.1073/pnas.94.11.5961 [12] DUFOUR D L. Cyanide content of cassava (Manihot esculenta, Euphorbiaceae) cultivars used by Tukanoan Indians in Northwest Amazonia [J]. Economic Botany, 1988, 42(2): 255 − 266. doi: 10.1007/BF02858929 [13] BRADSHAW J E. Root and tuber crops[M]. Berlin: Springer Science & Business Media, 2010: 46 − 61. [14] 时涛, 李超萍, 蔡吉苗, 等. 木薯新叶斑病病原鉴定及其生物学特性[J]. 热带作物学报, 2010, 31(3): 457 − 463. [15] 李超萍, 时涛, 刘先宝, 等. 国内木薯病害普查及细菌性萎蔫病安全性评估[J]. 热带作物学报, 2011, 32(1): 116 − 121. doi: 10.3969/j.issn.1000-2561.2011.01.024 [16] ZHU Z, SHI J, XU W, et al. Three ERF transcription factors from Chinese wild grapevine Vitis pseudoreticulata participate in different biotic and abiotic stress-responsive pathways [J]. Journal of Plant Physiology, 2013, 170: 923 − 933. doi: 10.1016/j.jplph.2013.01.017 [17] MUNOZ-BODNAR A, PEREZ-QUINTERO A L, GOMEZ-CANO F, et al. RNAseq analysis of cassava reveals similar plant responses upon infection with pathogenic and non-pathogenic strains of Xanthomonas axonopodis pv. manihotis [J]. Plant Cell Reports, 2014, 33(11): 1901 − 1912. doi: 10.1007/s00299-014-1667-7 [18] 杨帆, 苗灵凤, 胥晓, 等. 植物对干旱胁迫的响应研究进展[J]. 应用与环境生物学报, 2007, 13(4): 586 − 591. doi: 10.3321/j.issn:1006-687x.2007.04.031 [19] 金慧, 栾雨时. 转录因子在植物抗病基因工程中的研究进展[J]. 中国生物工程杂志, 2010, 30(10): 94 − 99. [20] FELTEN J, VAHALA J, LOVE J, et al. Ethylene signaling via Ethylene Response Factors (ERFs) modifies wood development in hybrid aspen [J]. BMC Proceedings. BioMed Central, 2011, 5(S7): I15. [21] FUJIMOTO S Y, OHTA M, USUI A, et al. Arabidopsis ethylene- responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression [J]. The Plant Cell, 2000, 12(3): 393 − 404. [22] ZHANG G, CHEN M, LI L, et al. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco [J]. Journal of Experimental Botany, 2009, 60(13): 3781 − 3796. doi: 10.1093/jxb/erp214 [23] BETHKE G, SCHEEL D, LEE J. Sometimes new results raise new questions: the question marks between mitogen-activated protein kinase and ethylene signaling [J]. Plant Signaling & Behavior, 2009, 4(7): 672 − 674. [24] ZAREI A, KORBES A P, YOUNESS P, et al. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis [J]. Plant Molecular Biology, 2011, 75(4/5): 321 − 331. -




 下载:
下载: