-
脱落酸(abscisic acid, ABA)是一种经典植物激素,在植物应对干旱、高盐和低温等非生物胁迫中起重要的调节作用。植物遭遇低温、干旱和高盐等逆境胁迫时,内源ABA迅速积累,应对环境胁迫的压力[1-2]。ABA诱导细胞中分子伴侣、LEA蛋白、防冻蛋白、糖、脯氨酸等保护剂的积累,或激活解毒机制,调节氧化还原平衡或改变离子转运来重建稳态,从而增强植物非生物胁迫的耐受性[3]。此外,ABA还调节植物种子成熟、种子休眠和萌发、营养发育、幼苗建立、根生长、开花、气孔运动和衰老等各方面的生长发育[4]。
人们对ABA信号传导途径已经有了较深入的了解。在高等植物逆境胁迫时,ABA含量增加,结合于受体PYR/PYL/RCAR蛋白内腔的配体结合位点,诱导受体蛋白的构象变化,ABA被封闭在受体蛋白内,重塑了受体蛋白的表面,为磷酸酶PP2Cs提供相互作用的结合位点,形成PYR/PYL/RCAR-ABA-PP2C复合物,从而阻断了底物进入PP2Cs的活性位点[5-7]。PP2Cs是ABA信号通路的负调节因子,可阻止ABA信号的传递,降低了植物对环境胁迫的耐受性[8-9]。PP2Cs与ABA受体的互作,增强了PYR/PYL/RCARs与ABA的结合力,其亲和力增加了大约10倍[10-11],从而减弱PP2C对下游蔗糖非发酵相关蛋白激酶2(Sucrose non-fermentating1-related protein kinase 2,SnRK2)的抑制作用[5-7]。一些没有与ABA结合的PYR/PYL/RCARs也可以自由与PP2Cs相互作用,但外源性ABA显著增强受体对PP2C活性的抑制作用[12]。在ABA信号转导途径中,SnRK2s是一个重要环节,能够磷酸化下游的转录因子和其他蛋白,从而调控植物的逆境反应。拟南芥有38个SnRKs,分为SnRK1、SnRK2和SnRK3三个亚家族[13],其中,SnRK2家族有10个成员。SnRK2s通过磷酸化bZIP23,介导干旱胁迫反应[14];通过磷酸化ABI5,介导ABA调控种子萌发[15];通过磷酸化ICE1,介导ABA调控抗寒性的形成[16]。低温胁迫下,拟南芥内源ABA含量增加,SnRK2.6/OST1被激活,从而磷酸化下游ICE1,增强它的转录活性和稳定性,提高了植株的抗寒性[16]。又因为JA也调节ICE1的转录[17],所以SnRK2.6/OST1在ABA和JA信号途径交互调控植物的ICE-CBF途径中起重要作用。
巴西橡胶树(Hevea brasiliensis Muell. Arg.)(简称橡胶树)原产南美洲亚马逊河流域,是商业用天然橡胶的主要来源。我国的橡胶树种植区位于世界热带北缘,经常性遭遇的寒流是我国发展橡胶树种植业面临的一种主要的非生物逆境。所以选育“高产抗寒”品种一直是我国橡胶树育种的首要目标。通过60余年的杂交育种,选育出一批抗寒性强的橡胶树品种,为研究橡胶树低温胁迫抗性形成机制提供了优良材料。橡胶树无性系‘93-114’是我国自主选育的次生代抗寒品种,尤其是抗平流型寒害的能力强。笔者从橡胶树无性系‘93-114’的叶片中分离到一个SnRK2基因家族的一个成员—HbSnRK2.7。为了探索蛋白激酶基因HbSnRK2.7在巴西橡胶树应对低温胁迫反应中的作用,采用q-PCR技术,分析了该基因在外源ABA处理和低温胁迫下的表达模式,以及不同抗寒性种质间的表达差异,旨在为进一步鉴定 HbSnRK2.7的功能提供理论依据。
-
实验材料为处于一蓬叶稳定期的橡胶树抗寒种质‘桂研73-165’、‘GT1’、‘INA873’、‘湛试32-713’、‘93-114’和不抗寒种质‘PB235’、‘热垦514’、‘热垦515’、‘海垦1’、‘热垦501’的芽接苗,该材料于2020年芽接于中国热带农业科学院种苗培育基地。多糖多酚植物总RNA提取试剂盒(离心柱型)和FastKing cDNA第一链合成试剂盒(去基因组)(天根生化科技有限公司);荧光定量所需酶(2×TB GreenTM Premix Ex TaqTMⅡ)(大连宝生物公司);其他生化试剂为进口或国产分析纯试剂;基因扩增和荧光定量引物(生工生物工程上海股份有限公司)。
-
低温处理恒温培养箱(PGR15,COVVILN,加拿大)条件设置为:光照强度为125 μmol·m−2·s−1,光照周期16 h/8 h,温度为28 ℃,相对湿度为80%。将生长一致且健康的30株芽接苗置于培养箱,恢复培养2 d,平均分为处理组和对照组,然后将处理组幼苗置于4 ℃的恒温培养箱中,其他设置条件不变,分别在0 、2 、4 、8、12、24 h时间点收集叶片,每个时间点选择15株幼苗,每株幼苗采集1片树叶,等量5片树叶制成1个混合样,每个时间点制成3个混合样,对照同样采集样品。用0.1 mmol·L−1 ABA 喷施整株幼苗,然后置于28 ℃的恒温培养箱中继续培养,分别在0、2、4、8、12和24 h时间点收集叶片,每个时间点选择15株幼苗,每株幼苗采集1片树叶,等量5片树叶制成1个混合样,每个时间点制成3个混合样[18]。ABA处理和低温处理共用对照。采集的样品迅速置于液氮中,用于提取总RNA。分别采集一蓬叶稳定期的橡胶树种质‘桂研73-165’、‘GT1’、‘INA873’、‘湛试32-713’、‘93-114’、‘PB235’、‘热垦514’、‘热垦515’、‘海垦1’、‘热垦501’芽接苗的树皮,每个种质采集5株幼苗制成1个混合样品,3个生物学重复,迅速置于液氮中,用于提取总RNA。
-
按照多糖多酚植物总RNA提取试剂盒说明书提取样品的总RNA。按照cDNA合成试剂盒的说明书合成cDNA。然后置于−20 ℃冰箱中保存备用。
-
从转录组[19]中,筛选1个有差异表达的 Unigene,注释为SnRK2家族成员,通过橡胶树基因组数据库[20],获得该基因cDNA序列。借助NCBI比对分析,确定该基因为HbSnRK2.7,并获得完整的基因开放阅读框序列。依据HbSnRK2.7的开放阅读框序列,设计引物(表1),扩增基因开放阅读框全长。利用 RT-PCR 技术,从橡胶树‘93-114’叶片中扩增出基因HbSnRK2.7的开放阅读框全长。RT-PCR扩增体系为 50 μL,高保真酶 25 μL、引物HbSnRK2.7-FL-F和HbSnRK2.7-FL-R各1 μL、模板1 μL、ddH2O添加至50 μL。扩增程序:95 ℃预变性5 min;95 ℃变性10 s,58 ℃退火30 s,72 ℃延伸2 min,35个循环;72 ℃再延伸5 min。然后1%琼脂糖凝胶电泳,胶回收,连接转化,挑阳性菌落,送广州艾基生物技术公司进行测序验证。
表 1 橡胶树基因HbSnRK2.7扩增引物和荧光定量引物
基因名称 引物用途 引物名称 引物序列 HbSnRK2.7 基因扩增 HbSnRK2.7-FL-F ATGGATCGATCGACGATTACTG HbSnRK2.7-FL-R TTATTGCAATGCATAAACTATCTC HbSnRK2.7 荧光定量 HbSnRK2.7-Q-F GAACTTCGGGGTTGCCAGAC HbSnRK2.7-Q-R TGCCTGAGGGACCTGTGATT HbActin7a HbActin7a-Q-F GGCACTTTGGTACTCAAGTC HbActin7a-Q-R GAAGCATCCCAATCACTCTC HbRH8 HbRH8-Q-F TCACAGGGTTGGTAGATCAG HbRH8-Q-R CCAAGCTCTTGCTCAATCC -
通过DNAMAN V9软件对HbSnRK2.7进行多重序列比对、基因序列翻译、蛋白质分析等方面的分析。通过Expasy软件对HbSnRK2.7蛋白的分子量和等电点进行分析。通过MEGA5.10软件对HbSnRK2.7的氨基酸序列进行进化树分析。通过NetPhos 3.1软件分析HbSnRK2.7的磷酸化位点。
-
利用CFX384实时荧光定量PCR系统(Bio-Rad,加利福尼亚,美国)进行q-PCR实验。依据基因丰度,将cDNA稀释10倍作为q-PCR模板。实验反应体系和方法参照文献 [18]。内参基因为HbActin7a和HbRH8。基因HbSnRK2.7荧光定量引物和内参引物见表1。HbSnRK2.7相对表达量通过2−△△Cq 法进行分析[21]。
-
采用t-test方法对基因相对表达量进行差异显著性分析(P<0.05为显著性差异;P<0.01为极显著差异)。
-
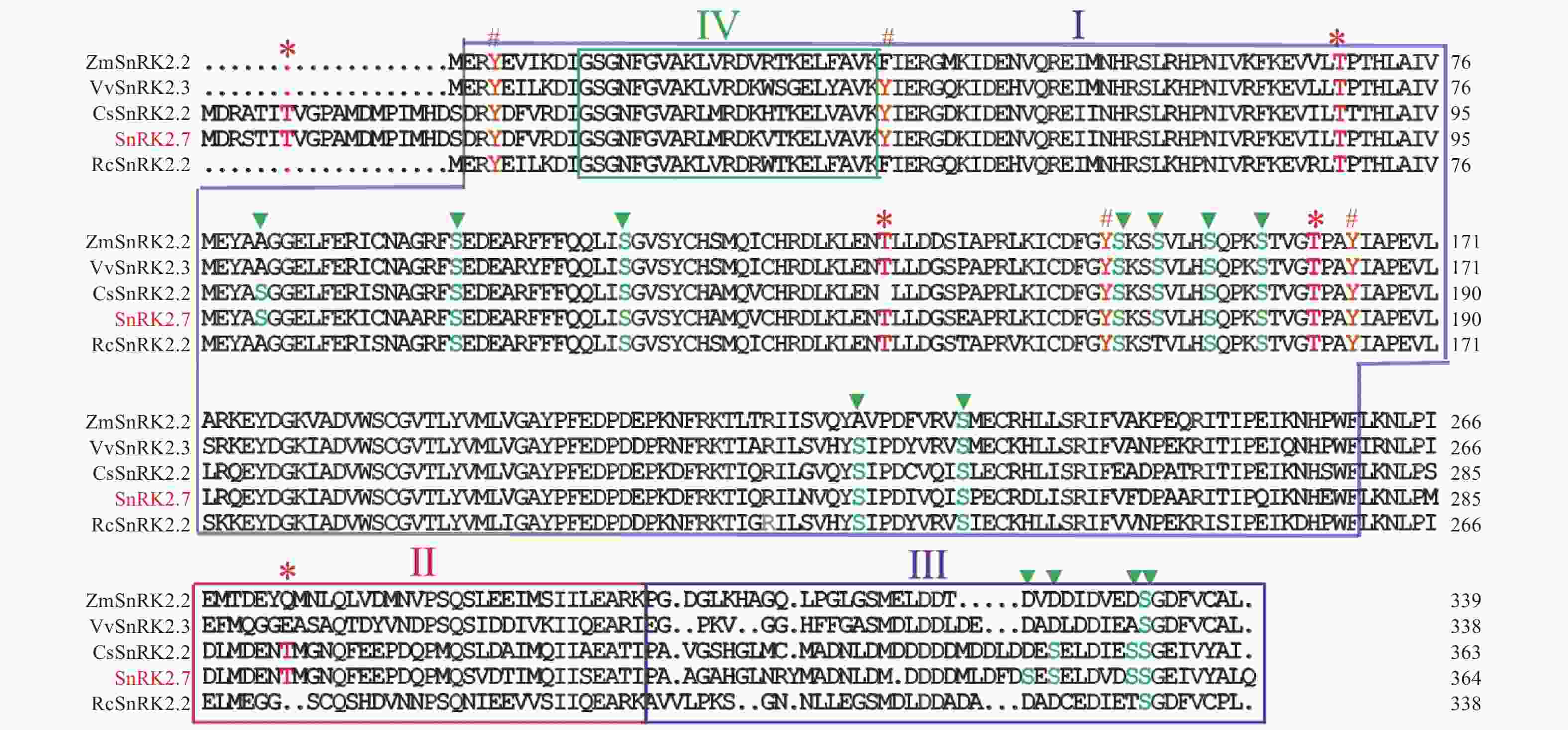
在HbSnRK2.7阅读框的两端设计引物,通过RT-PCR技术及核苷酸测序技术,以橡胶树‘93-114’的cDNA为模板,克隆了HbSnRK2.7基因的完整阅读框(图1)。HbSnRK2.7包含364个氨基酸,预测的分子量和等电点分别为41.39 kD和4.70。HbSnRK2.7与拟南芥(Arabidopsis thaliana L Heynh.)AtSnRK2.2、AtSnRK2.3、黄瓜(Cucumis sativus L.)CsSnRK2.2、葡萄(Vitis vinifera L)VvSnRK2.3的核苷酸序列的同源性分别为73.39%、74.98%、80.87%和77.99%。HbSnRK2.7氨基酸序列与黄瓜CsSnRK2.2、葡萄VvSnRK2.3、蓖麻(Ricinus communis L)RcSnRK2.2、玉米(Zea mays L)ZmSnRK2.2的氨基酸序列同源性分别为90.96%, 84.07%, 65.11%和67.58%。虽然氨基酸序列同源性有一定差异,但是它们具有保守的丝氨酸/苏氨酸蛋白激酶结构域(结构域Ⅰ),可催化磷酸基从ATP转移到蛋白质底物上的丝氨酸/苏氨酸残基上(图2)。另外,它们都有结构域Ⅱ、结构域Ⅲ和结构域Ⅳ。结构域Ⅱ是HbSnRK2.7响应非生物胁迫所需的结构域;结构域Ⅲ紧挨结构域Ⅱ,位于蛋白的C端,是ABA依赖的HbSnRK2.7激活所需结构域;结构域Ⅳ位于结构域Ⅰ内,HbSnRK2.7蛋白的N端,是与ATP结合的保守结构域(图2)。结构域的保守性表明它们可能具有相似的生物学功能。通过NetPhos 3.1软件分析HbSnRK2.7的磷酸化位点,结果显示HbSnRK2.7含有13个丝氨酸、5个苏氨酸和4个酪氨酸磷酸化位点,大部分磷酸化位点集中在丝氨酸/苏氨酸蛋白激酶结构域(图2)。聚类结果显示,橡胶树HbSnRK2.7与拟南芥AtSnRK2.2、AtSnRK2.3、黄瓜CsSnRK2.2、葡萄VvSnRK2.3等属于SnRK2亚家族的第Ⅲ组(图3)。
-
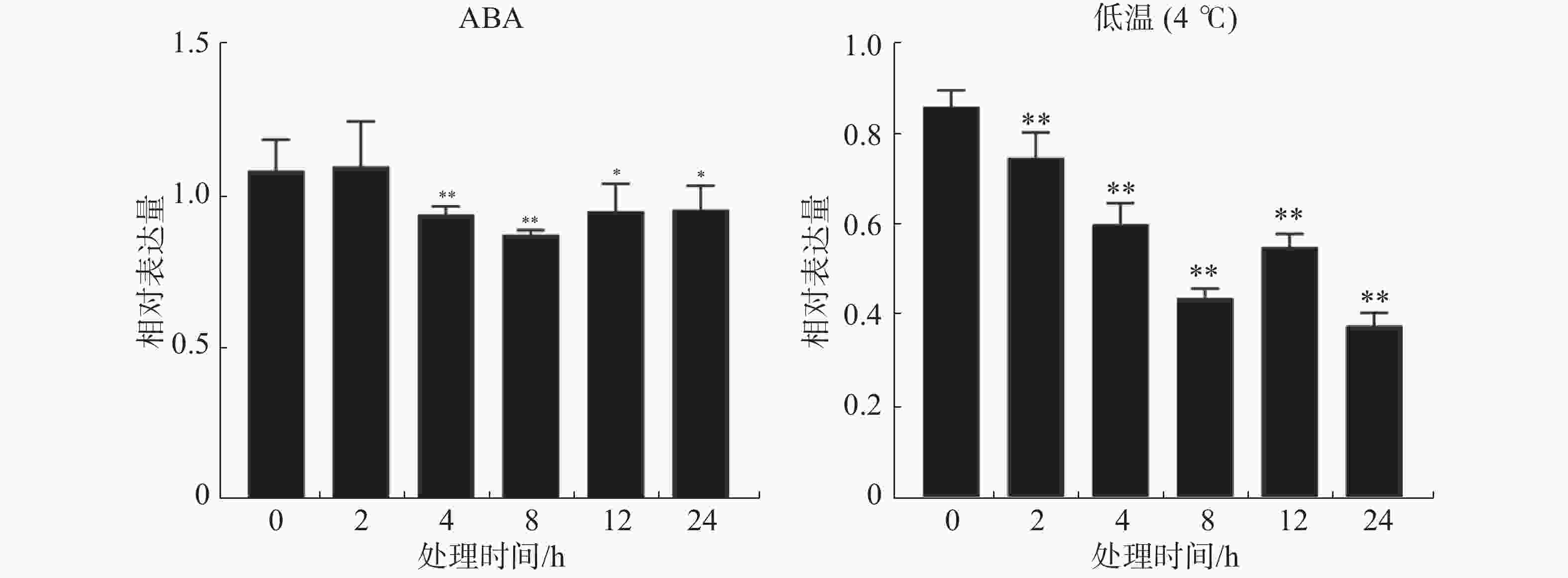
ABA处理下,抗寒橡胶树‘93-114’树皮中的HbSnRK2.7基因呈下调表达趋势。在ABA处理2 h时,HbSnRK2.7的表达量稍微变动,没有显著差异。处理4 h时,HbSnRK2.7已极显著下调表达,在处理8 h时,表达量下调到最低点。在处理12 h和24 h时,HbSnRK2.7表达量稍微上调且趋于稳定,但是其表达量仍显著低于ABA处理0 h时的表达量。低温胁迫下,HbSnRK2.7基因在橡胶树‘93-114’中的表达呈下调趋势。在低温胁迫2 h时,HbSnRK2.7的表达量极显著降低。低温胁迫4 h和8 h时,HbSnRK2.7继续下调表达,在低温胁迫12 h时,表达量上调,但极显著低于低温胁迫0 h的表达量。低温胁迫24 h时,HbSnRK2.7表达量下调到最低,是低温胁迫0 h的表达量的1/2(图4)。
-
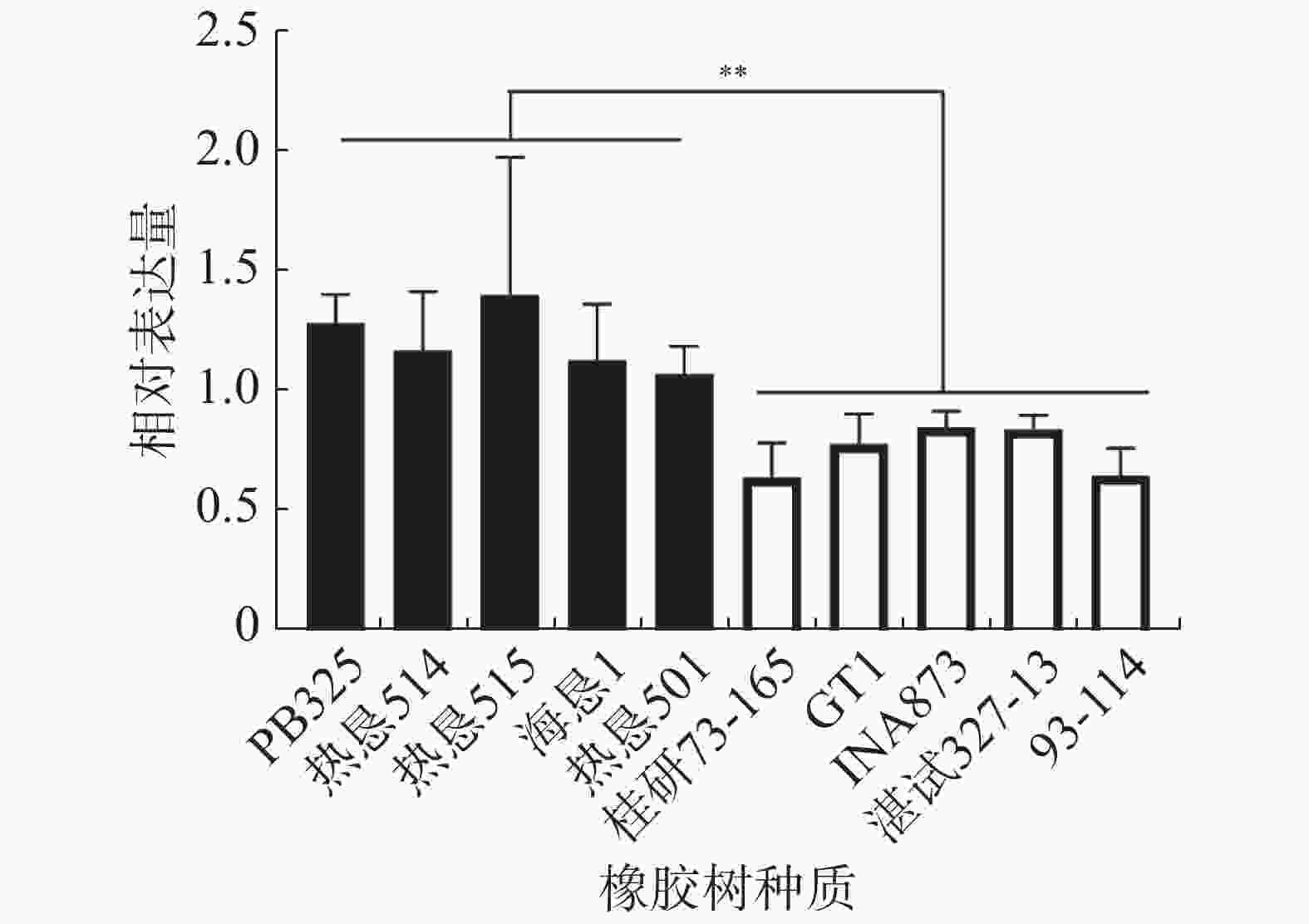
橡胶树HbSnRK2.7在不抗寒种质‘热垦515’中的本底表达最高,在抗寒种质‘桂研73-165’中的本底表达最低。在不抗寒种质‘PB235’、‘热垦514’、‘热垦515’、‘海垦1’和‘热垦501’中的整体表达量显著高于抗寒种质‘桂研73-165’、‘GT1’、‘INA873’、‘湛试32-713’和‘93-114’中的表达量(图5)。
-
本研究利用RT-PCR技术从橡胶树‘93-114’中分离鉴定了丝氨酸/苏氨酸蛋白激酶HbSnRK2.7基因,橡胶树HbSnRK2.7基因与草本植物拟南芥AtSnRK2.2、AtSnRK2.3、黄瓜CsSnRK2.2、木本植物葡萄VvSnRK2.3的同源性较高,核苷酸序列的一致性在73%以上,氨基酸序列的一致性在65%以上。它们具有共同的保守丝氨酸/苏氨酸蛋白激酶催化结构域[22-24],同源性比较高,该结构域是SnRK2蛋白磷酸化功能的核心催化中心,其中包含ATP结合结构域,可催化磷酸基从ATP转移到蛋白质底物上的丝氨酸/苏氨酸残基上,使底物磷酸化[25]。SnRK2s参与了植物对非生物胁迫和脱落酸(ABA)依赖的植物发育的响应。SnRK2亚家族分为3个组:组Ⅰ中的蛋白激酶是不被ABA激活的;组Ⅱ中的蛋白激酶不被ABA激活或被ABA微弱激活,取决于植物种类;组Ⅲ中的蛋白激酶强烈响应ABA诱导[22]。HbSnRK2.7属于丝氨酸/苏氨酸蛋白激酶家族的SnRK2亚家族的组Ⅲ成员,具有ABA激活所需结构域,推测HbSnRK2.7能够响应ABA的刺激反应。另外,HbSnRK2.7还具有响应非生物胁迫所需的结构域,推测HbSnRK2.7能够响应非生物胁迫反应。磷酸化位点主要集中在HbSnRK2.7的丝氨酸/苏氨酸蛋白激酶催化结构域,磷酸化/去磷酸化调控了HbSnRK2.7对底物的磷酸化活性。
植物在遭受干旱、低温、高盐等环境胁迫时,其内源激素ABA迅速合成和积累,在植物应对环境胁迫中起着重要作用[1-2]。SnRK2s蛋白激酶是ABA信号途径的关键环节,介导依赖ABA的非生物胁迫反应[26-27]。拟南芥AtSnRK2.2、AtSnRK2.3响应ABA刺激反应,呈下调表达趋势[28-30]。HbSnRK2.7与黄瓜CsSnRK2.2核苷酸序列的同源性为80.87%,CsSnRK2.2响应ABA诱导下调表达[23]。HbSnRK2.7与葡萄VvSnRK2.1核苷酸序列的同源性为75.08%,VvSnRK2.1同样响应ABA诱导下调表达[31]。HbSnRK2.7与拟南芥AtSnRK2.2、AtSnRK2.3、黄瓜CsSnRK2.2、葡萄VvSnRK2.1的表达模式一致,受ABA诱导下调表达。推测HbSnRK2.7可能在ABA信号转导途径中起着负调控的作用。然而与HbSnRK2.7同属于亚家族Ⅲ的水稻SAPK8、SAPK9和SAPK10受ABA诱导上调表达[32],暗示即使同一亚家族的成员,在不同的物种对于ABA刺激的响应也存在差异。
SnRK2s是ABA信号途径的重要环节,参与植物低温胁迫反应[33]。拟南芥AtSnRK2.6受低温胁迫上调表达,激活的AtSnRK2.6磷酸化ICEs,干扰了E3连接酶HOS1与ICEs的互作,从而抑制了ICEs经26S蛋白酶体的降解,增强其稳定性和转录活性,提高植物的抗寒性[16]。橡胶树HbSnRK2.6A/B/C受ABA和低温诱导上调表达,且通过调控抗寒关键因子HbICE2的转录活性,参与ABA调控橡胶树响应低温胁迫[34]。本研究克隆的橡胶树HbSnRK2.7对ABA处理和低温胁迫的响应与HbSnRK2.6s相反,该基因既在ABA处理下显著下调表达,又受低温胁迫显著下调表达,而且在不抗寒种质中的表达量显著高于抗寒种质,表明橡胶树HbSnRK2.7可能作为负调控因子,介导ABA依赖的橡胶树低温胁迫抗性的形成。与HbSnRK2.7同源性比较高的玉米ZmSnRK2.2[35]、葡萄VvSnRK2.1、VvSnRK2.3 [36]在低温胁迫下呈下调表达模式。鹅掌楸(Liriodendron chinense)SnRK2家族2个成员(Lchi00543和Lchi01348)受ABA和低温诱导同样下调表达[37]。HbSnRK2.7类似的基因都是对应物种SnRK2家族的极少数成员,与其他受ABA和低温正向调控的多数成员不同[38]。因此,这些基因可能作为负调控因子在植物响应低温胁迫中起着特殊的作用。
橡胶树蛋白激酶基因SnRK2.7的克隆及表达
DOI: 10.15886/j.cnki.rdswxb.20230004
 CSTR: 32425.14.j.cnki.rdswxb.20230004
CSTR: 32425.14.j.cnki.rdswxb.20230004
Cloning and expression analysis of sucrose non-fermenting 1-related protein kinase 2.7 (SnRK2.7) gene in Hevea brasiliensis
-
摘要: 为了探索蔗糖非发酵相关蛋白激酶2(Sucrose non-fermentating1-related protein kinase 2,SnRK2)在巴西橡胶树响应低温胁迫应答中的作用,采用RT-PCR技术,从橡胶树抗寒无性系93-114的叶片中克隆了SnRK2亚家族的一个成员,命名为HbSnRK2.7。该基因包含一个1 095 bp的开放阅读框,编码364个氨基酸。HbSnRK2.7的分子量为41.39 kD,等电点为4.70,具有保守丝氨酸/苏氨酸蛋白激酶结构域,预测其能催化磷酸基从ATP转移到蛋白质底物上的丝氨酸/苏氨酸残基上,也是磷酸化位点的集中区域,还具有非生物胁迫所需的结构域和ABA(脱落酸)依赖的HbSnRK2.7激活所需结构域。q-PCR结果显示,HbSnRK2.7基因响应ABA诱导,呈下调表达模式。在ABA处理8 h时,表达量下调到最低点。在低温胁迫下,HbSnRK2.7的表达量极显著降低。低温胁迫4 h和8 h时,HbSnRK2.7持续下调表达,低温胁迫24 h时,HbSnRK2.7表达量下调到最低,大约为非胁迫表达量的1/2。而且,HbSnRK2.7在不抗寒种质中的表达量显著高于抗寒种质。这些结果表明,HbSnRK2.7可能作为负调控因子介导橡胶树依赖ABA的低温胁迫抗性形成。
-
关键词:
- 巴西橡胶树 /
- 蔗糖非发酵相关蛋白激酶 /
- 荧光定量PCR /
- 低温胁迫抗性 /
- 脱落酸信号途径
Abstract: To explore the role of sucrose non-fermentation associated protein kinase 2 (SnRK2) in the response of Hevea brasiliensis to low temperature stress, a member of the SnRK2 subfamily, named HbSnRK2.7, was cloned from the leaves of the cold-resistant clone Reyan ‘93-114’ of Hevea brasiliensis by using RT-PCR. The gene cloned contains an open reading frame of 1 095 bp that encodes a protein of 364 amino acids. HbSnRK2.7 has a conserved serine/threonine protein kinase domain with a molecular weight of 41.39 kD and an isoelectric point of 4.70. It is predicted that it can catalyze the transfer of phosphate groups from ATP to the serine/threonine residues on the protein substrate, which is also the concentrated region of phosphorylation sites. It also has domains needed for abiotic stress and ABA-dependent domains for HbSnRK2.7 activation. Quantitative PCR (qPCR) results showed that HbSnRK2.7 gene was down-regulated in response to ABA induction. When treated with ABA for 8 h, the expression was down-regulated to the lowest point. Under low temperature stress, the expression of HbSnRK2.7 decreased significantly. The expression of HbSnRK2.7 was down-regulated continuously under low temperature stress for 4 h and 8 h, and the expression of HbSnRK2.7 was down-regulated to the lowest level at 24 h under low temperature stress, which was about 1/2 of non-stress expression. Moreover, the expression of HbSnRK2.7 was significantly higher in the cold-susceptible clones than in the cold-resistant clones. These results suggest that HbSnRK2.7 may act as a negative regulatory factor to mediate the formation of low temperature stress resistance of Hevea brasiliensis dependent on ABA.-
Key words:
- Hevea brasiliensis Muell. Arg /
- SnRK2 /
- q-PCR /
- cold resistance /
- ABA signaling pathway
-
图 3 橡胶树HbSnRK2.7蛋白与其他植物的SnRK2蛋白的系统进化树分析
橡胶树HbSnRK2.7(ATO58465)、黄瓜CsSnRK2.2(XP_004148038)、拟南芥AtSnRK2.1(NP_196476)、AtSnRK2.2(NP_001190047)、AtSnRK2.3(NP_001318893)、AtSnRK2.4(NP_001031021)、AtSnRK2.5(NP_201170)、AtSnRK2.6 (NP_567945)、AtSnRK2.7(NP_195711)、AtSnRK2.8(NP_001077839)、AtSnRK2.9(NP_179885)、AtSnRK2.10(NP_176290)、水稻OsSAPK10(NP_001389116)、OsSAPK9(NP_001391667)、OsSAPK8(NP_001389202)、OsSAPK3(XP_052134301)、OsSAPK2A(NP_001390090)、OsSAPK1(NP_001389097)、OsSAPK6(NP_001388802)、OsSAPK5(NP_001389390)、OsSAPK4(NP_001383791)、OsSAPK7(BAH93169)、葡萄VvSnRK2.3(XP_002284959)、VvSnRK2.2(XP_002269221)、VvSnRK2.5(XP_003634478)、VvSnRK2.7(XP_002267922)、VvSnRK2.8(XP_002262731)。
表 1 橡胶树基因HbSnRK2.7扩增引物和荧光定量引物
基因名称 引物用途 引物名称 引物序列 HbSnRK2.7 基因扩增 HbSnRK2.7-FL-F ATGGATCGATCGACGATTACTG HbSnRK2.7-FL-R TTATTGCAATGCATAAACTATCTC HbSnRK2.7 荧光定量 HbSnRK2.7-Q-F GAACTTCGGGGTTGCCAGAC HbSnRK2.7-Q-R TGCCTGAGGGACCTGTGATT HbActin7a HbActin7a-Q-F GGCACTTTGGTACTCAAGTC HbActin7a-Q-R GAAGCATCCCAATCACTCTC HbRH8 HbRH8-Q-F TCACAGGGTTGGTAGATCAG HbRH8-Q-R CCAAGCTCTTGCTCAATCC -
[1] YAMAGUCHI-SHINOZAKI K, SHINOZAKI K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses[J]. Annual Review of Plant Biology, 2006, 57: 781 − 803. doi: 10.1146/annurev.arplant.57.032905.105444 [2] QIN F, SHINOZAKI K, YAMAGUCHI-SHINOZAKI K. Achievements and challenges in understanding plant abiotic stress responses and tolerance[J]. Plant and Cell Physiology, 2011, 52(9): 1569 − 1582. doi: 10.1093/pcp/pcr106 [3] KLINE K G, BARRETT-WILT G A, SUSSMAN M R. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid[J]. Proceedings of the National Academy of Sciences of theUnited States of America, 2010, 107(36): 15986 − 15991. doi: 10.1073/pnas.1007879107 [4] FINKELSTEIN R. Abscisic Acid synthesis and response[J]. The Arabidopsis Book, 2013, 11: e0166. doi: 10.1199/tab.0166 [5] MELCHER K, NG L M, ZHOU X E, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors[J]. Nature, 2009, 462(7273): 602 − 608. doi: 10.1038/nature08613 [6] MIYAZONO K I, MIYAKAWA T, SAWANO Y, et al. Structural basis of abscisic acid signalling[J]. Nature, 2009, 462(7273): 609 − 614. doi: 10.1038/nature08583 [7] YIN P, FAN H, HAO Q, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins[J]. Nature Structural & Molecular Biology, 2009, 16(12): 1230 − 1236. doi: 10.1038/nsmb.1730 [8] HAIDER M S, ZHANG C, KURJOGI M M, et al. Insights into grapevine defense response against drought as revealed by biochemical, physiological and RNA-Seq analysis[J]. Scientific Reports, 2017, 7(1): 13134. doi: 10.1038/s41598-017-13464-3 [9] SHAZADEE H, KHAN N, WANG J, et al. Identification and expression profiling of protein phosphatases (PP2C) gene family in Gossypium hirsutum L[J]. International Journal of Molecular Sciences, 2019, 20(6): 1395. doi: 10.3390/ijms20061395 [10] MA Y, SZOSTKIEWICZ I, KORTE A, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors[J]. Science, 2009, 324(5930): 1064 − 1068. doi: 10.1126/science.1172408 [11] SANTIAGO J, DUPEUX F, ROUND A, et al. The abscisic acid receptor PYR1 in complex with abscisic acid[J]. Nature, 2009, 462(7273): 665 − 668. doi: 10.1038/nature08591 [12] HAO Q, YIN P, LI W, et al. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins[J]. Molecular Cell, 2011, 42(5): 662 − 672. doi: 10.1016/j.molcel.2011.05.011 [13] HRABAK E M, CHAN C W M, GRIBSKOV M, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases[J]. Plant Physiology, 2003, 132(2): 666 − 680. doi: 10.1104/pp.102.011999 [14] ZONG W, TANG N, YANG J, et al. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes[J]. Plant Physiology, 2016, 171(4): 2810 − 2825. doi: 10.1104/pp.16.00469 [15] WANG Z, REN Z, CHENG C, et al. Counteraction of ABA-mediated inhibition of seed germination and seedling establishment by ABA signaling terminator in Arabidopsis[J]. Molecular Plant, 2020, 13(9): 1284 − 1297. doi: 10.1016/j.molp.2020.06.011 [16] DING Y, LI H, ZHANG X, et al. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis[J]. Developmental Cell, 2015, 32(3): 278 − 289. doi: 10.1016/j.devcel.2014.12.023 [17] HU Y, JIANG L, WANG F, et al. Jasmonate regulates the inducer of cbf expression-C- repeat binding factor/dre binding factor1 cascade and freezing tolerance in Arabidopsis[J]. The Plant Cell, 2013, 25(8): 2907 − 2924. doi: 10.1105/tpc.113.112631 [18] 李言, 余文才, 陈月异, 等. 巴西橡胶树过氧化氢酶基因HbCAT2的克隆及表达分析[J]. 分子植物育种, 2019, 17(21): 6993 − 7002. [19] DENG X, WANG J, LI Y, et al. Comparative transcriptome analysis reveals phytohormone signalings, heat shock module and ROS scavenger mediate the cold-tolerance of rubber tree[J]. Scientific Reports, 2018, 8(1): 4931. doi: 10.1038/s41598-018-23094-y [20] TANG C, YANG M, FANG Y, et al. The rubber tree genome reveals new insights into rubber production and species adaptation[J]. Nature Plants, 2016, 2(6): 16073. doi: 10.1038/nplants.2016.73 [21] LI Y, QUAN C, YANG S, et al. Functional identification of ICE transcription factors in rubber tree[J]. Forests, 2022, 13(1): 52. doi: 10.3390/f13010052 [22] KULIK A, WAWER I, KRZYWIŃSKA E, et al. SnRK2 protein kinases-key regulators of plant response to abiotic stresses[J]. Omics, 2011, 15(12): 859 − 872. doi: 10.1089/omi.2011.0091 [23] WAN Z, LUO S, ZHANG Z, et al. Identification and expression profile analysis of the SnRK2 gene family in cucumber[J]. PeerJ, 2022, 10: e13994. doi: 10.7717/peerj.13994 [24] BONEH U, BITON I, SCHWARTZ A, et al. Characterization of the ABA signal transduction pathway in Vitis vinifera[J]. Plant Science, 2012, 187: 89 − 96. doi: 10.1016/j.plantsci.2012.01.015 [25] LIN Z, LI Y, WANG Y, et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling[J]. Nature Communications, 2021, 12(1): 2456. doi: 10.1038/s41467-021-22812-x [26] BELDA-PALAZóN B, ADAMO M, VALERIO C, et al. A dual function of SnRK2 kinases in the regulation of SnRK1 and plant growth[J]. Nature Plants, 2020, 6(11): 1345 − 1353. doi: 10.1038/s41477-020-00778-w [27] WAADT R, SELLER C A, HSU P K, et al. Plant hormone regulation of abiotic stress responses[J]. Nature Reviews Molecular Cell Biology, 2022, 23(10): 680 − 694. doi: 10.1038/s41580-022-00479-6 [28] BOUDSOCQ M, BARBIER-BRYGOO H, LAURIÈRE C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana[J]. The Journal of Biological Chemistry, 2004, 279(40): 41758 − 41766. doi: 10.1074/jbc.M405259200 [29] FUJII H, VERSLUES P E, ZHU J K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis[J]. The Plant Cell, 2007, 19(2): 485 − 494. doi: 10.1105/tpc.106.048538 [30] KOBAYASHI Y, MURATA M, MINAMI H, et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors[J]. The Plant Journal, 2005, 44(6): 939 − 949. doi: 10.1111/j.1365-313X.2005.02583.x [31] 陈娜娜. 葡萄SnRK2基因家族的全基因组鉴定、表达分析及VvSnRK2.2基因的功能验证 [D]. 南京: 南京农业大学, 2013. [32] KOBAYASHI Y, YAMAMOTO S, MINAMI H, et al. Differential activation of the rice sucrose nonfermenting1-related protein kinase 2 family by hyperosmotic stress and abscisic acid[J]. Plant Cell, 2004, 16(5): 1163 − 1177. doi: 10.1105/tpc.019943 [33] CHEN X, DING Y, YANG Y, et al. Protein kinases in plant responses to drought, salt, and cold stress[J]. Journal of Integrative Plant Biology, 2021, 63(1): 53 − 78. doi: 10.1111/jipb.13061 [34] WANG X, LIU W C, ZENG X W, et al. HbSnRK2.6 functions in ABA-regulated cold stress response by promoting HbICE2 transcriptional activity in Hevea brasiliensis. Int J Mol Sci. , 2021, 22(23): 12707. [35] HUAI J, WANG M, HE J, et al. Cloning and characterization of the SnRK2 gene family from Zea mays[J]. Plant Cell Reports, 2008, 27(12): 1861 − 1868. doi: 10.1007/s00299-008-0608-8 [36] 马宗桓, 毛娟, 李文芳, 等. 葡萄SnRK2家族基因的鉴定与表达分析[J]. 园艺学报, 2016, 43(10): 1891 − 1902. [37] HUSSAIN Q, ZHENG M, CHANG W, et al. Genome-wide identification and expression analysis of SnRK2 gene family in dormant vegetative buds of Liriodendron chinense in response to abscisic acid, chilling, and photoperiod. Genes, 2022, 13(8): 1305. [38] MASZKOWSKA J, SZYMAŃSKA K P, KASZTELAN A, et al. The multifaceted regulation of SnRK2 kinases[J]. Cells, 2021, 10(9): 2180. -




 下载:
下载: