-
鸟巢蕨(Asplenium nidus)是铁角蕨科Aspleniaceae巢蕨属的一个种,又名山苏花,是热带雨林大型附生植物[1]。原生于亚洲东南部、澳大利亚东部、印度尼西亚、印度和非洲东部等地[2-3]。《中国植物志》记载我国有鸟巢蕨分布于台湾、福建、香港、广东、广西、海南、湖南、四川、贵州、云南和西藏等省份和地区。鸟巢蕨植物不仅可以丰富森林生物多样性,还可调节森林生态系统的水分与养分循环,且具有药用、食用与园林绿化价值。近年来,由于人为活动干扰,野生鸟巢蕨被大量采集,生境缩小且破碎化严重,导致其遗传多样性降低。加上种间杂交严重,导致遗传背景错综复杂,已影响到其品种改良、新品种培育、保护和开发利用。因此,加强对鸟巢蕨遗传多样性与进化研究对鸟巢蕨种质资源保护和利用以及育种和遗传改良具有重要意义。SRAP(sequence-related amplified polymorphism)是相关序列扩增多态性的简称,利用双引物设计对基因的ORFS(Open Reading Frames,开放阅读框)进行区域扩增是该技术特点,因不同物种内含子、启动子与间隔区的长度不同而产生多态性[4-6]。SRAP技术具有操作方便、多态性丰富、重复性好、易于测序、速度快和成本低等特性,被广泛运用于动物、植物、微生物的品种鉴定、遗传图谱构建、遗传多样性检测、基因定位和比较基因组学等研究[7-8]。SRAP标记通过直接分析遗传物质多态性作为不同种群划分依据更为可行和客观,能够较好地开展遗传多样性分析,可避免因季节性生理生化差异和不同发育阶段而影响指标的准确性[9-10]。因此,可通过分子标记技术分析鸟巢蕨亲缘关系,得到更加科学和稳定的分析结果。海南岛是鸟巢蕨分布中心之一[11-12],野生种群集中分布于霸王岭、俄贤岭、黎母山和五指山等热带雨林区域。不同种群间的鸟巢蕨在叶形、株高、孢子量、巢基大小方面存在一定差异[13-14]。为揭示鸟巢蕨自然种群状况,促进野生鸟巢蕨资源有效保护和利用,本研究采用SRAP标记技术,对海南岛霸王岭、俄贤岭、黎母山和五指山6个鸟巢蕨自然种群进行遗传多样性分析,旨在探明海南岛鸟巢蕨种群之间遗传关系、亲缘关系,进而揭示海南岛不同地理环境对鸟巢蕨遗传多样性及种群分化的影响,为鸟巢蕨保育和遗传资源利用提供理论依据。
HTML
-
于2011年5月−8月及2019年3月−4月,在海南岛霸王岭、俄贤岭、黎母山及五指山6个自然种群进行采样,每个种群范围均>2 km2,6个自然种群相互间地理距离>10 km。每个采集种群内采样个体间间隔距离>1 m。共采集96份植株叶片样本,硅胶干燥后,带回实验室,提取DNA。各种群地理位置、生境及采样个体数详见表1。
种群
Population地点
Location生境
Habitat样本数/份
Sampling/No.经度(E)
Longitude纬度(N)
Latitude海拔/m
AltitudeBW 霸王岭 (Bawangling) 雨林沟谷 石生、附生 16 109°12′ 19°06′ 755~770 EXL 俄贤岭 (Exianling) 雨林沟谷 石生、附生 16 109°38′ 18°53′ 900~1000 LMS1 黎母山 (Limushan 1) 雨林沟谷 石生、附生 8 109°42′ 19°10′ 740~760 LMS2 黎母山 (Limushan 2) 雨林沟谷 石生、附生 7 109°25′ 19°26′ 720~740 LMS3 黎母山 (Limushan 3) 雨林沟谷 石生、附生 23 109°18′ 19°38′ 700~750 WZS 五指山 (Wuzhishan) 雨林沟谷 石生、附生 26 109°41′ 18°54′ 800~900 Table 1. Location, habitat, and sample size of the 6 populations of Asplenium nidus in Hainan Island
-
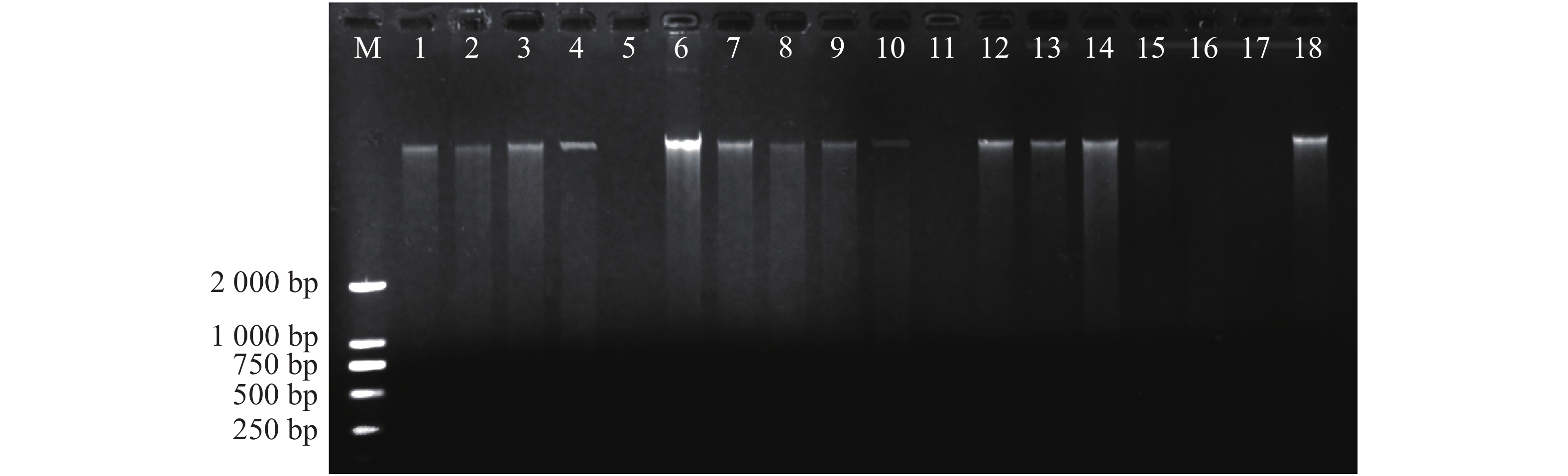
采用北京天根生物有限公司生产的新型植物基因组DNA 提取试剂盒(DP305)提取鸟巢蕨叶片DNA,用w=1.0%琼脂糖凝胶电泳检测,图1为鸟巢蕨种群基因组DNA电泳图。提取样品基因组DNA 用核酸微量测定仪稀释至一致的质量浓度(50 mg·L−1),置于−20 ℃下保存备用。
-
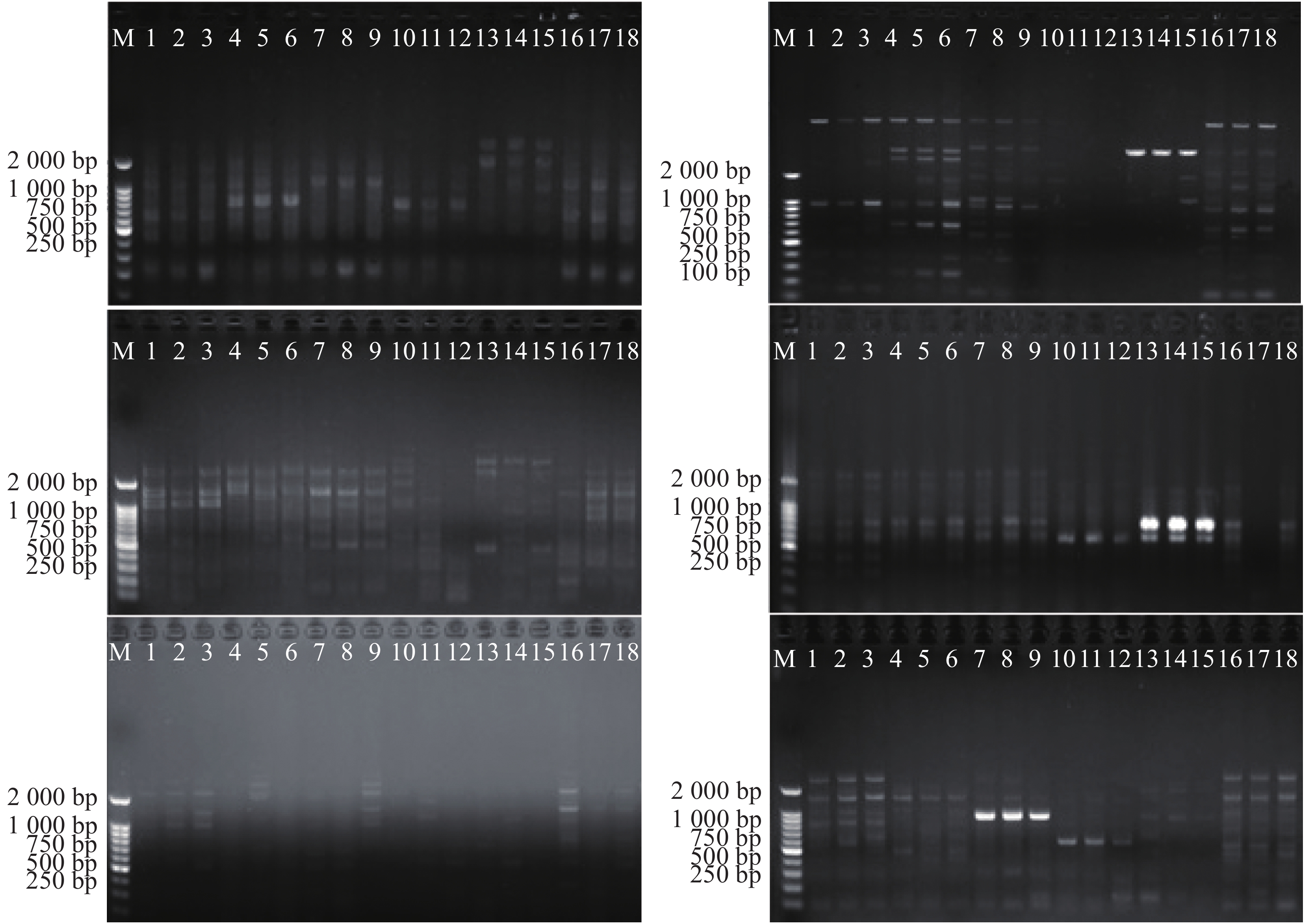
根据LI等[15]的方法,设计正向引物17个,反向引物20个,由北京三博生物技术有限公司合成。PCR扩增反应使用TC-512PCR仪(Techne,Ltd.)。PCR扩增体系:1U的Taq DNA酶,1.5 mmol·L−1的Mg2+,0.20 mmol·L−1的dNTP(Fermentas Biotec.)和25 ng的DNA,0.5 μmol·L−1的引物。PCR扩增程序:94 ℃预变性5 min, 94 ℃变性30 sec,52 ℃退火30 sec,72 ℃延伸1 min,共35个循环;最后72 ℃终延伸10 min,4 ℃保存[16]。扩增产物在w=6%变性聚丙烯酰胺凝胶上(65 w恒定功率)电泳2~2.5 h。电泳完毕后,进行银染,通过成像仪拍照并保存。
-
将SRAP记录的电泳图进行人工读带,由图1可得,以同一位点有带记作“1”,无带记作“0”,仅记录清晰、稳定且长度在300~2 500 bp的扩增条带,形成0-1矩阵二元数据输入电脑。
采用POPGENE32软件衡量SRAP水平上遗传变异程度,对全部种群和各个种群分别进行遗传参数分析,分别对有效等位基因数(Ne, effective number of allele)、等位基因观察值(Na,number of allele)、多态位点百分率(PPB,percentage of polymorphic bands)、Nei’s基因多样性(He,Nei’s genetic diversity)、Shannon信息指数(Ho,Shannon’s information index)[16-17]、种群总基因多样度(Ht,total gene diversity)、种群内基因多样度(Hs,gene diversity within populations)、种群间的基因流(N,geneflow),种群间遗传分化系数(Gst,coefficient of population differentiation)、Nei’s无偏差遗传距离(D,Nei’s genetic distance)和遗传一致度(I,genetic identity)等参数进行计算[18]。
应用GenAlEx v.6(Genetic Analysis in Excel)软件[19]对种群内遗传变异和种群间遗传变异进行分子变异分析(AMOVA),计算遗传变异在种群内和种群间分布。Shannon信息指数HO由如下公式计算:
式中,HO为Shannon信息指数,Pi为第i 条带的表型频率。
遗传分化系数(coefficient of genetic differentiation,Gst)用来估算种群间的遗传分化程度,即种群间遗传多样性占总遗传多样性的比例。
式中,Gst为遗传分化系数,Ht为种群的遗传多样度,Hs为各种群内的基因多样度,m为该位点上的等位基因,n为居群总数,rj为该位点上第j个等位基因在总种群中的平均频率,qij为第i个种群在该位点上第j个等位基因频率,mi为第i个种群在该位点上的等位基因数。
基因流(gene flow)用来估算种群间基因交流程度,为每个世代在种群间迁移的个体数,是通过遗传分化系数来估算的:
式中,Nm为基因流,N为有效种群大小;m为种群每代迁移率;Gst为遗传分化系数。
根据 Nei’s 遗传距离,使用NTSYS-pc 2.1软件对种群进行UPGMA聚类分析,样本数为85。
1.1. 材料
1.2. DNA提取
1.3. SRAP扩增与检测分析
1.4. 数据统计分析
-
利用17个正向引物与20个反向引物共340对引物组合进行扩增,筛选出10对条带清晰、稳定性高、特异性好且多态性丰富的PCR扩增产物用于鸟巢蕨的遗传多样性分析[15]。其中,正向和反向引物序列见表2,10对引物组合为:ME3-EM3、ME4-EM10、ME5-EM8、ME6-EM16、ME5-EM7、ME10-EM10、ME14-EM6、ME16-EM14、ME16-EM10、ME3-EM4。
Forward Primer 5′−3′ Reverse Primer 5′−3′ ME1 TGAGTCCAAACCGGAGC EM1 GACTGCGTACGAATTAAT ME2 TGAGTCCAAACCGGATA EM2 GACTGCGTACGAATTTGC ME3 TGAGTCCAAACCGGAAT EM3 GACTGCGTACGAATTGAC ME4 TGAGTCCAAACCGGACC EM4 GACTGCGTACGAATTAAC ME5 TGAGTCCAAACCGGAAG EM5 GACTGCGTACGAATTTGA ME6 TGAGTCCAAACCGGTAA EM6 GACTGCGTACGAATTGCA ME7 TGAGTCCAAACCGGTGC EM7 GACTGCGTACGAATTGAG ME8 TGAGTCCAAACCGGTCC EM8 GACTGCGTACGAATTCTG ME9 TGAGTCCAAACCGGTAG EM9 GACTGCGTACGAATTGTC ME10 TGAGTCCAAACCGGTTG EM10 GACTGCGTACGAATTCAG ME11 TGAGTCCAAACCGGTGT EM11 GACTGCGTACGAATTCCA ME12 TGAGTCCAAACCGGTCA EM12 GACTGCGTACGAATTCAC ME13 TGAGTCCAAACCGGGAC EM13 GACTGCGTACGAATTGCC ME14 TGAGTCCAAACCGGGTA EM14 GACTGCGTACGAATTGGT ME15 TGAGTCCAAACCGGGGT EM15 GACTGCGTACGAATTCGG ME16 TGAGTCCAAACCGGCAG EM16 GACTGCGTACGAATTATG ME17 TGAGTCCAAACCGGCTA EM17 GACTGCGTACGAATTAGC EM18 GACTGCGTACGAATTACG EM19 GACTGCGTACGAATTTAG EM20 GACTGCGTACGAATTTCG Table 2. Forward and Reverse Primer sequences used in SRAP analysis
引物用于对海南岛鸟巢蕨6个野生种群共96个个体进行PCR扩增,由扩增结果(图2)可看出,一共扩增出184个清晰稳定的条带,筛选6条清晰条带进行分析,其中174条具有多态性,多态位点百分率为94.57%。从图1可看出,每个引物扩增出的条带为5~9条,平均条带数为6.9。条带分子量在250~2 000 bp之间,而表3里的各种群间多态位点百分率存在较大的差异。基于SRAP分子标记的鸟巢蕨物种水平多态位点百分比(PPB)为94.57%,SRAP检测到的种群多态位点百分比例(PPB)在76.09%(LMS2种群)~94.57%(LMS3种群)之间,平均为85.15%,鸟巢蕨种群间遗传变异程度较大(76.09%~94.57%),说明各种群均具有较高的遗传多样性水平。
种群
Population等位基因
观察值(No)
Observed number
of alleles有效等位
基因数(Ne)
Effective number
of allelesNei基因
多样性(He)
Nei’s genetic diversityShannon信息
指数(Ho)
Shannon’s information
index多态位点数
Number of
polymorphic loci多态位点
百分率(PPB)
Percentage of polymorphic
loci/%BW 1.8152±0.3892 1.4799±0.3509 0.2822±0.1814 0.4227±0.2510 150 81.52 EXL 1.8533±0.3548 1.5343±0.3491 0.3094±0.1714 0.4597±0.2381 157 85.33 LMS1 1.8533±0.3548 1.4901±0.3350 0.2919±0.1686 0.4403±0.2301 157 85.33 LMS2 1.7609±0.4277 1.5224±0.3771 0.2961±0.1933 0.4346±0.2696 140 76.09 LMS3 1.9457±0.2273 1.5332±0.2273 0.3225±0.1378 0.4881±0.1796 174 94.57 WZS 1.8804±0.3253 1.5087±0.3198 0.3040±0.1607 0.4577±0.2191 162 88.04 Population level 1.8515±0.3465 1.5114±0.3265 0.3010±0.1689 0.4505±0.2313 156.7 85.15 Species level 1.9457±0.2273 1.5893±0.2864 0.3475±0.1329 0.5181±0.1728 174 94.57 Table 3. Genetic diversity (Mean±SE) of 6 natural populations of Asplenium nidus based on SRAP markers
-
POPGENE软件分析结果表明,种群间和种群内均存在一定遗传变异,但主要发生在种群内部。种群总基因多样度(Ht)为0.346 2±0.018 3,其中,种群内基因多样度(Hs)为0.301 0±0.014 2,即86.94%的遗传分化发生在种群内部;种群间遗传分化系数(Gst)为0.1306,即种群间遗传分化为13.06%,说明鸟巢蕨种群间遗传分化水平较高,且遗传变异主要发生在种群内部。种群间基因流Nm=1.664 2,表明鸟巢蕨之间基因流动畅通。
AMOVA分析表明,遗传变异以居群内变异为主,此分析结果与POPGENE的分析结果基本一致,由表4可看出,显著的遗传分化(P = 0.010)于种群内部和种群间均存在,遗传变异在种群间占总遗传变异的10.62%,在种群内占89.53%。
变异来源
Source of variance自由度
df平方和
SS均方
MS变异大小
Variancecomponents变异百分率/%
Percentageof variation概率
P种群间 5 471.146 94.229 3.959 10.62% 0.010 种群内 90 2 997.989 33.311 33.311 89.53% 0.010 总计 95 3 469.135 37.270 100% Table 4. Analysis of molecular variance (AMOVA) of natural populations of Asplenium nidus
-
从表5可知,鸟巢蕨6个种群的平均遗传相似度为0.922 4,平均遗传距离为0.080 9。其中,BW和EXL的遗传距离最近,为0.050 8,LMS2和LMS3遗传距离最远,为 0.116 6。
种群 Population BW EXL LMS1 LMS2 LMS3 WZS BW **** 0.950 5 0.925 6 0.926 9 0.897 7 0.923 7 EXL 0.050 8 **** 0.942 1 0.929 5 0.911 5 0.934 5 LMS1 0.077 3 0.059 7 **** 0.915 0 0.937 6 0.928 9 LMS2 0.076 0 0.073 1 0.088 8 **** 0.889 9 0.897 6 LMS3 0.107 9 0.092 7 0.064 4 0.116 6 **** 0.925 5 WZS 0.079 4 0.067 7 0.073 8 0.108 0 0.077 4 **** 注:种群同表1一致。上部为遗传相似度, 下部为遗传距离。
Note:The population is consistent with Table 1. The upper part is genetic similarity and the lower part is genetic distance.Table 5. Genetic similarity and genetic distance among the 6 populations of Asplenium nidus based on SRAP
SRAP 标记对6个种群鸟巢蕨野外种群遗传距离的UPGMA聚类分析结果表明,海南岛6个鸟巢蕨种群可分为三大类:由图3可得,地理位置较近的BW和EXL为第I类;同一地区的LMS1和LMS3聚类在了一起为第II类;而LMS2与其余种群为第Ⅲ类,其遗传距离最大,遗传分化也最显著。
2.1. 海南岛鸟巢蕨自然种群的遗传多样性
2.2. 鸟巢蕨的遗传分化
2.3. 鸟巢蕨种群间的遗传距离与相似系数
-
遗传多样性是生物多样性重要组成部分,是生态系统多样性和物种多样性的基础[16-17],也是分析物种进化潜力和未来命运重要方向[18]。遗传多样性大小是物种长期进化的产物,是其生存适应和发展进化的前提[20]。遗传多样性越高的物种对环境变化适应能力就越强,越容易在其分布范围内扩展和开拓新生境[20-21]。
本研究结果表明,海南岛野生鸟巢蕨种群具有较高遗传多样性,多态位点百分比(PPB)在76.09%(LMS2种群)~94.57%(LMS3种群)之间,平均为85.15%。该结果与潘丽芹等[22]的荷叶铁线蕨自然居群的遗传多样性研究和董元火等[23]的水蕨的生境及其遗传多样性分析结果相似,但与黄庆阳等[24]对香鳞毛蕨种质资源遗传多样性的AFLP分析研究结果存在一定差异。原因可能是研究材料地理分布差异较大,或使用的分子标记技术不同所导致。本研究中,野生鸟巢蕨种群间生境差异不大,但随着海拔增加,气候、土壤、植被等也随之发生了明显变化,因而造成6个种群较高遗传多样性。其中,LMS3地区遗传多样性最高(Ne、He、Ho分别为1.533 2、0.322 5、0.488 1),可能黎母山(LMS)作为海南三大江河(南渡江、万泉河、昌化江)的发源地,石灰岩沟谷较多,符合鸟巢蕨对生境要求。加上黎母山林区属热带季风气候,季风带来长期潮湿更有利于鸟巢蕨孢子传播和更新,而霸王岭样地鸟巢蕨种群遗传多样性最小(Ne、He、Ho分别为1.479 9、0.282 2、0.422 7);有两个方面的原因:一是所采集个体的海拔分布较低,受人为干扰较重;另一原因可能是鸟巢蕨分布地受季风影响较小,不利于孢子传播和附着。本研究结果表明,在海南岛6个鸟巢蕨种群中LMS3鸟巢蕨种群内部遗传变异最多,而BWL鸟巢蕨种群内部遗传变异则相对较少。
-
遗传结构指种群中遗传变异分布的时空格局,其在基因流、基因突变和自然选择的共同作用下形成,也与物种进化史和生物学特性有关[25-26]。
鸟巢蕨虽然在种群和物种水平均具有较高的遗传多样性,但相比较而言,种群水平遗传多样性要低于物种水平,与种群间具有较高基因流(Nm=1.664 2,Nm>1)有关,使漂变成为划分种群遗传结构主导因素,表明鸟巢蕨距离较远种群间也存在一定程度基因交流[27-28]。遗传分化程度Gst为0.130 6,在0.05~0.15之间,属于中等程度遗传分化。
基于SRAP标记得出6个鸟巢蕨野生种群多态位点百分率、Shannon信息指数(Ho)、Nei’s基因多样性(He)、种群内遗传多样性指数和总遗传多样性指数,通过AMOVA分析结果显示总的种群间变异为11%,说明鸟巢蕨种群内存在丰富的遗传变异,6个鸟巢蕨野生分布种群间均有较高遗传变异,黎母山种群与其他5个种群遗传变异最明显。蒋速飞等 [29]发现,丰富的遗传变异可能是物种具有较高遗传多样性的原因之一 。由于鸟巢蕨是孢子繁殖,孢子不易于长距离迁徙,易造成地理隔离,推测地理隔离可能是造成鸟巢蕨种群间遗传分化较大原因,而较高遗传变异可能是本研究中各鸟巢蕨种群间遗传多样性较高的原因之一[30]。
-
物种遗传多样性水平能反映其适应能力和进化潜力,还有助于物种稀有或濒危原因及过程的探讨[31]。对珍稀濒危物种保护方针和措施的制定,如采样策略迁地或就地保护选样等都有赖于人们对物种遗传多样性认识[32-33]。鸟巢蕨靠孢子繁殖,而孢子适宜在潮湿的环境生长,海南岛气候十分适宜鸟巢蕨繁殖生长,本研究也证实海南岛鸟巢蕨具较高遗传多样性,说明在本研究所选区域现阶段人为活动对鸟巢蕨的数量和遗传多样性影响较小,各区域还有一定数量鸟巢蕨种群分布。
值得注意的是,一旦生境遭到破坏会直接导致孢子不能传播和存活,鸟巢蕨原生基因就会丧失,其遗传多样性也会随之降低。建议鸟巢蕨保护应以就地保护为主,可选择鸟巢蕨种群遗传多样性较高的黎母山保护区区域(如本研究的黎母山LMS3区域)建立级别更高的自然保护区,加大保护和自然恢复力度,扩大保护边界。针对鸟巢蕨孢子不易于长距离迁徙导致不同种群之间遗传分化较大这一现象,可对不同自然群体中鸟巢蕨个体迁地保护,结合采用重引入技术进行自然居群恢复,提高种群竞争力和多样性,扩大群体间基因交流,最大限度保障鸟巢蕨遗传多样性,促进热带雨林国家公园建成。









 DownLoad:
DownLoad: