-
单锌指DNA 结合蛋白(DNA binding with one finger,Dof)是一类植物特异性转录因子,由多基因家族编码。Dof蛋白大约由200~400个氨基酸残基(Amino acid, aa)组成,Dof蛋白含有2个主要的保守结构域:即N末端具有高度保守DNA结合域和位于C末端的转录调控域[1]。Dof蛋白N−末端的DNA结合域由52个保守的氨基酸残基组成的CX2CX21CX2C单锌指结构,基序中的4个Cys残基和1个Zn2+共价结合,Dof蛋白的DNA结合域与不同植物的启动子DNA结合具有特异性,识别AAAG或互补序列CTTT基序作为核心序列元件[2],但是南瓜Dof蛋白AOBP为例外,AOBP蛋白特异识别AGTA序列[3]。位于C−末端的转录调控结构域的氨基酸序列不具有保守性,导致Dof蛋白在植物生长发育过程中的功能的多样性。自从第一个Dof(ZmDof1)基因在玉米中克隆以来[4],迄今为止从单细胞藻类到高等植物,越来越多的Dof基因被克隆鉴定或从基因组数据库中被预测出来。基于植物Gene bank数据库已被鉴定的Dof基因家族数目为拟南芥36个[5]、水稻30个[5]、玉米46个[6]、小麦31个[7]、大豆28个[8]、高粱28个[9]、马铃薯35个[10-11]、番茄34个[12]、菊花20个[13]、大白菜76个[14]、香蕉74个[15]、榴莲24个[16]、木薯45个[17]、辣椒33个[18]、葡萄25个[19]等,但目前关于荔枝Dof基因家族的系统分析尚未见报道。本研究利用课题组妃子笑荔枝果肉不同发育时期的转录组测序数据,采用生物信息学方法在转录组水平上对Dof基因家族进行系统鉴定,通过对Dof基因家族基本理化性质、保守结构域、系统进化和基因表达等进行分析,为进一步了解荔枝Dof基因家族(LcDof)的功能提供理论参考。
HTML
-
妃子笑荔枝果肉材料取自中国热带农业科学院南亚热带作物研究所荔枝种植园。分别取妃子笑果肉发育的3个时期(花后35 ,56,70 d)的样品,3个生物学重复,迅速置于液氮中,用于后续实验。
-
转录组测序委托广州基迪奥生物公司,利用Illumina HiSeqTM4000平台进行测序,对原始测序数据进行过滤、去接头和低质量值数据后,用reads组装软件Trinity 软件进行序列组装并获得Unigene 序列,共拼接组装获得61648个Unigenes。从植物转录因子数据库下载Dof转录因子序列作为探针,对本地妃子笑荔枝果肉转录组数据进行搜索,筛选出LcDof并利用SMART和Pfam 验证,删除不完整的基因。
-
分别采用ProtParam、Plant-mPlc Sever、MEME和SMART在线软件对Dof基因家族的相应基本理化性质、亚细胞定位、保守基序、基序功能注释进行生物信息学分析。MEME参数设置:基序最大值设为15,基序长度范围为6~100个氨基酸,其他参数为默认值。将荔枝、拟南芥和水稻中的Dof氨基酸序列通过Clustal X 1.83进行多重序列比对,进一步通过MEGA6.0邻接法(neighbor joining,NJ)将比对结果构建系统发育树。进化树参数设置:P-距离(P-distance)模型,成对删除(pairwise deletion)空位选项,Bootstrap method取值1 000。
-
Dof基因家族的表达数据基于课题组前期构建的“妃子笑”果肉发育RNA-seq数据库中的表达数据的rpkm值,利用Heml热图软件将Dof基因家族的表达数据均一化处理后制作热图。
1.1. 材料
1.2. LcDof鉴定与分析
1.3. LcDof生物信息学分析
1.4. LcDof基因家族在不同处理花穗发育过程中的表达特征分析
-
以拟南芥和水稻的Dof基因家族序列为探针,采用本地blast方法对荔枝果实发育RNA-seq数据库进行比对检索,同时利用Dof为关键词在RNA-seq数据库直接搜索;将得到的数据结果进一步进行整合分析,去除冗余序列,得到20个Dof蛋白序列。随后采用SMART和Pfam在线软件对Dof蛋白序列进行保守结构域分析,最终获得19个Dof蛋白序列(表1)。Dof基因家族成员编号按照Dof序列在RNA-seq数据库中的Unigene ID号出现顺序编号(表1)。LcDof蛋白序列长度范围为157~497 aa,平均长度为330.68 aa,LcDof18蛋白序列最短,LcDof15蛋白序列最长;LcDof蛋白序列的分子量(MW)为17.70~54.35 kDa,平均分子量为35.95 kDa。LcDof等电点(pI)在4.49~9.42,19个Dof蛋白中有6个等电点小于7显酸性;13个等电点大于7显碱性;LcDof蛋白平均等电点大于7,表明LcDof为弱碱性,在碱性的亚细胞环境中发挥作用。分析LcDof家族不稳定指数发现,LcDof4/9/11/14蛋白不稳定指数<40,为稳定蛋白,其余均为不稳定蛋白。分析LcDof家族脂肪族氨基酸指数发现,Dof家族的脂肪族氨基酸指数分布在46.2~66.32范围,脂肪族氨基酸指数与蛋白的热稳定性相关,表明Dof家族蛋白质间的热稳定性存在差异。LcDof家族蛋白质疏水性指数(Grand average of hydropathicity,GRAVY)均<0,表明LcDof家族蛋白均为亲水性蛋白。亚细胞定位预测结果表明,LcDof蛋白均定位于细胞核,这与转录因子定位于细胞核结果相符。
基因
GeneUnigene ID Dof
domain
结构域蛋白序列长度
Amino acids
length/aa分子量
Moleculer
weight/kDa等电点
PI不稳定指数
Instability
index脂肪族氨基酸
指数
Aliphatic index疏水指数
GRAVY亚细胞定位
Subcellular
localizationLcDof1 Unigene0010345 50~108 349 38.24 8.73 61.67 58.4 −0.63 细胞核 Nucleus LcDof2 Unigene0013981 118~176 469 51.02 6.33 60.6 57.63 −0.787 Nucleus. LcDof3 Unigene0014508 21~79 284 32.22 4.49 49.83 60 −0.624 Nucleus. LcDof4 Unigene0015472 37~95 214 21.82 4.61 26.91 55.14 −0.341 Nucleus. LcDof5 Unigene0019917 43~101 289 31.91 8.69 51.85 59.72 −0.75 Nucleus. LcDof6 Unigene0020262 29~87 336 36.73 7.18 49.53 50.21 −0.882 Nucleus. LcDof7 Unigene0022097 27~85 302 33.30 8.3 42.76 55.23 −0.719 Nucleus. LcDof8 Unigene0023140 47~105 316 34.87 8.79 41.45 54.91 −0.734 Nucleus. LcDof9 Unigene0025055 26~84 264 27.17 8.44 36.47 53.52 −0.376 Nucleus. LcDof10 Unigene0025382 69~127 325 35.09 9.32 64.41 64.25 −0.617 Nucleus. LcDof11 Unigene0027651 47~105 310 34.22 6.35 39.5 53.16 −0.669 Nucleus. LcDof12 Unigene0027696 21~79 274 30.13 9.26 47.09 52.26 −0.756 Nucleus. LcDof13 Unigene0032463 98~156 495 53.63 5.66 43.82 65.37 −0.526 Nucleus. LcDof14 Unigene0033612 18~76 237 24.82 8.45 35.97 49.83 −0.64 Nucleus. LcDof15 Unigene0033960 135~193 497 54.35 6.06 55.32 46.2 −0.923 Nucleus. LcDof16 Unigene0034259 146~204 495 54.33 8.27 49.68 53.8 −0.824 Nucleus. LcDof17 Unigene0050967 69~127 340 36.11 8.93 53.65 66.32 −0.513 Nucleus. LcDof18 Unigene0059627 40~98 157 17.70 9.42 47.12 50.25 −0.896 Nucleus. LcDof19 Unigene0060175 85~143 330 35.37 9.34 53.43 57.58 −0.632 Nucleus. Table 1. Litchi Dof gene family information
-
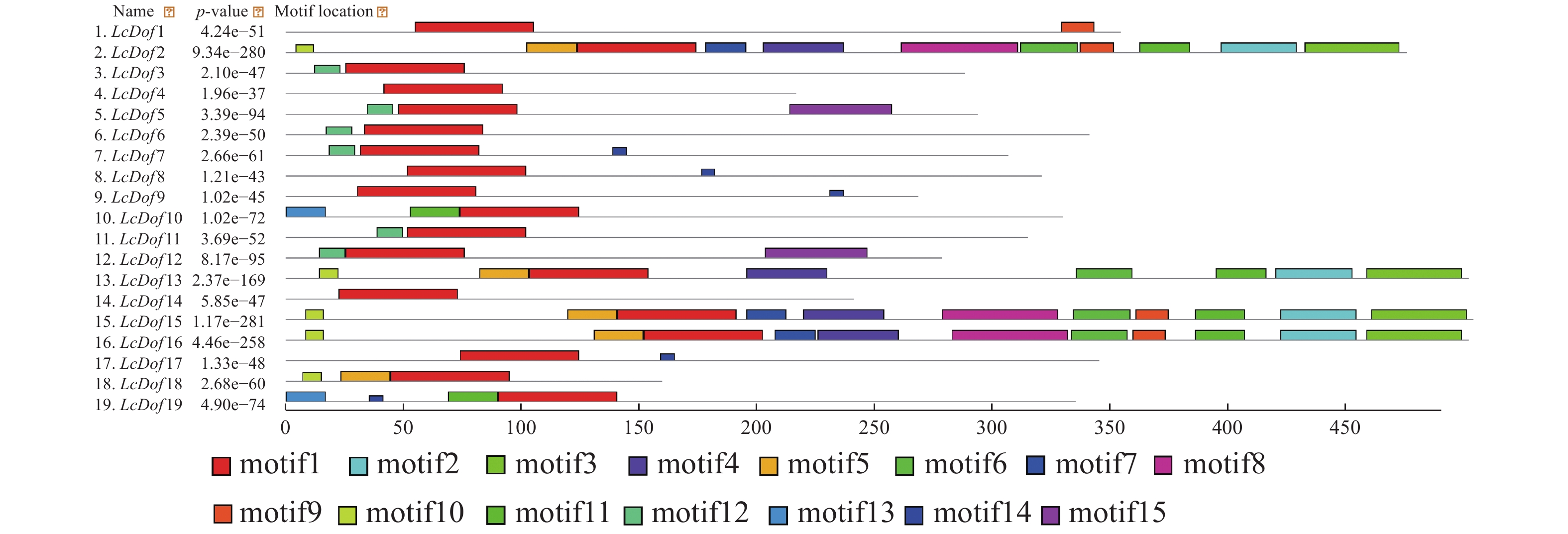
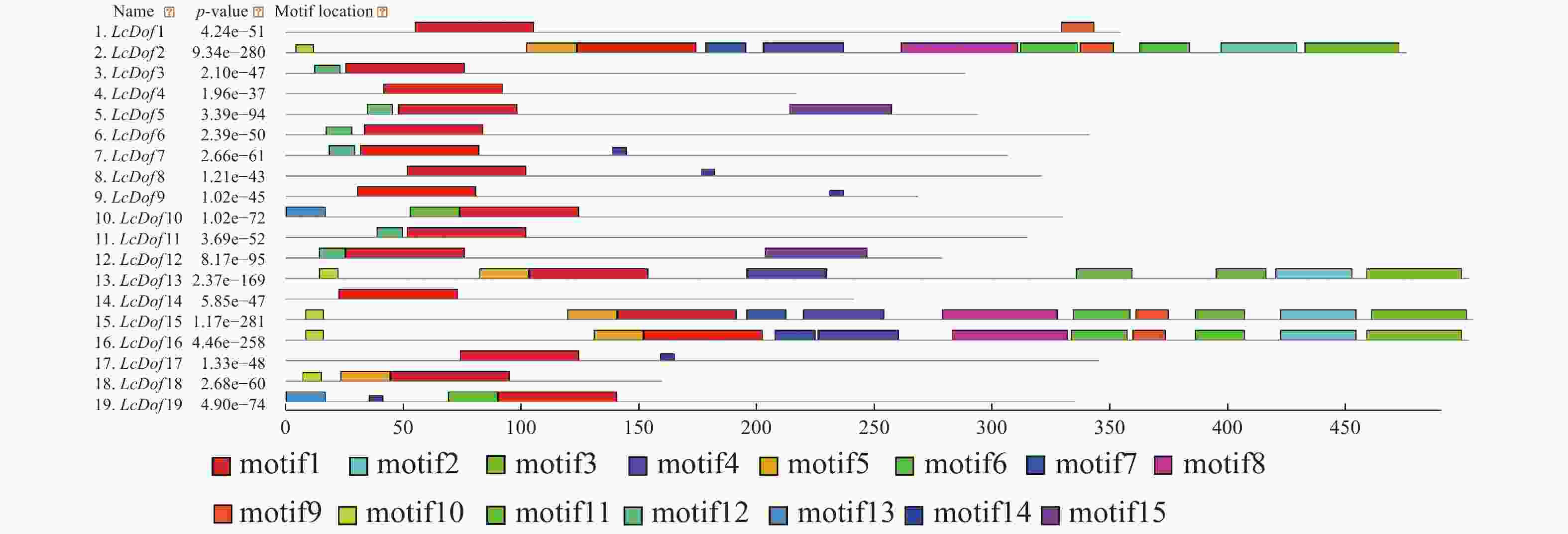
为了进一步了解 LcDof蛋白结构特征,利用MEME在线软件分析19个LcDof蛋白的保守基序,不同保守结构域在LcDof的位置如图1所示。LcDof基因家族中含有15个保守基序,并将得到的15个保守基序进一步进行功能注释(表2)的结果表明:15个基序中基序1出现在所有的蛋白质中,为N−末端高度保守的锌指结构域(zinc-finger Dof domain,zf-Dof),基序8、15为低密度复杂区,其余的12个基序没有对应的注释,功能未知。虽然荔枝Dof基因家族成员均含有基序1,但是LcDof基因家族成员之间包含的保守基序数目及种类存在一定的差异,其中LcDof2、LcDof15和LcDof16含有的基序数目最多,均含有11个保守基序;LcDof13含有8个基序;LcDof3、LcDof6和LcDof11均含2个基序(motif 1和motif12);LcDof8、LcDof9和LcDof17均含2个基序(motif 1和motif14)。LcDof4仅含有1个基序。在进化树中关系较近的LcDof成员间有类似的保守基序,如GroupIV中的LcDof2、LcDof15和LcDof16。LcDof中保守结构域组成相似的成员可能具有相近的基因功能。
基序
Motif基序长度/bp
Motif length基序序列
Motif sequence功能注释
Function annotation1 50 CPRCBSTNTKFCYYNNYNLSQPRHFCKTCRRYWTKGGTLRNVPVGGGCRK zf-Dof 2 32 ERCVLVPKTLRIDDPDEAAKSSIWATLGIKND 未知 3 40 GGGLFKGFQPKSDEKNRIAETSPVLQANPAALSRSLNFHE 未知 4 34 HHPSLKSNGTVLSFGSDAPLCDSMASVLNLADKK 未知 5 21 EQSESSESQEKTLKKPDKIJP 未知 6 24 YPWNPPVPPPAFCPPGFPMPFYPA 未知 7 17 AAHYRHITISEALQTAR 未知 8 49 ENGDDHSNGSSVTVSNSKEEGGKTAMQEPLMQNYQGFPPQIPCFPGPPW low complexity
低密度复杂区9 14 YWGCTIPGSWNMPA 未知 10 8 IKLFGKTI 未知 11 21 PGSGPNSPTLGKHSRDENALK 未知 12 11 ERKLRPQKEQA 未知 13 17 MVFPSVPLYLDPPNWQQ 未知 14 6 FDHHHH 未知 15 43 FPLQDFKPTLNFSJDGLGNGFGSLNGVQENGTGRLFFPFEELK low complexity
低密度复杂区Table 2. LcDof protein conserved motifs and functional annotations
-
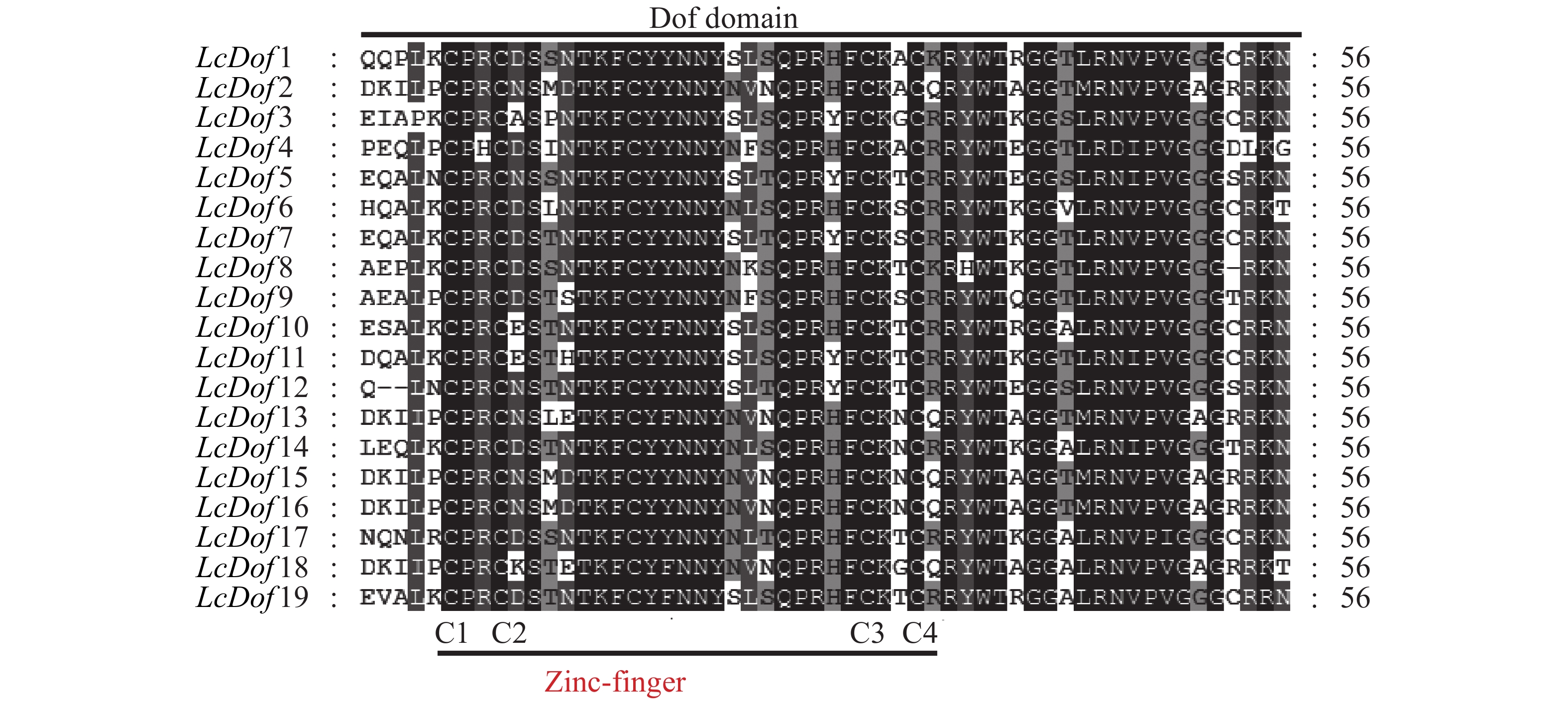
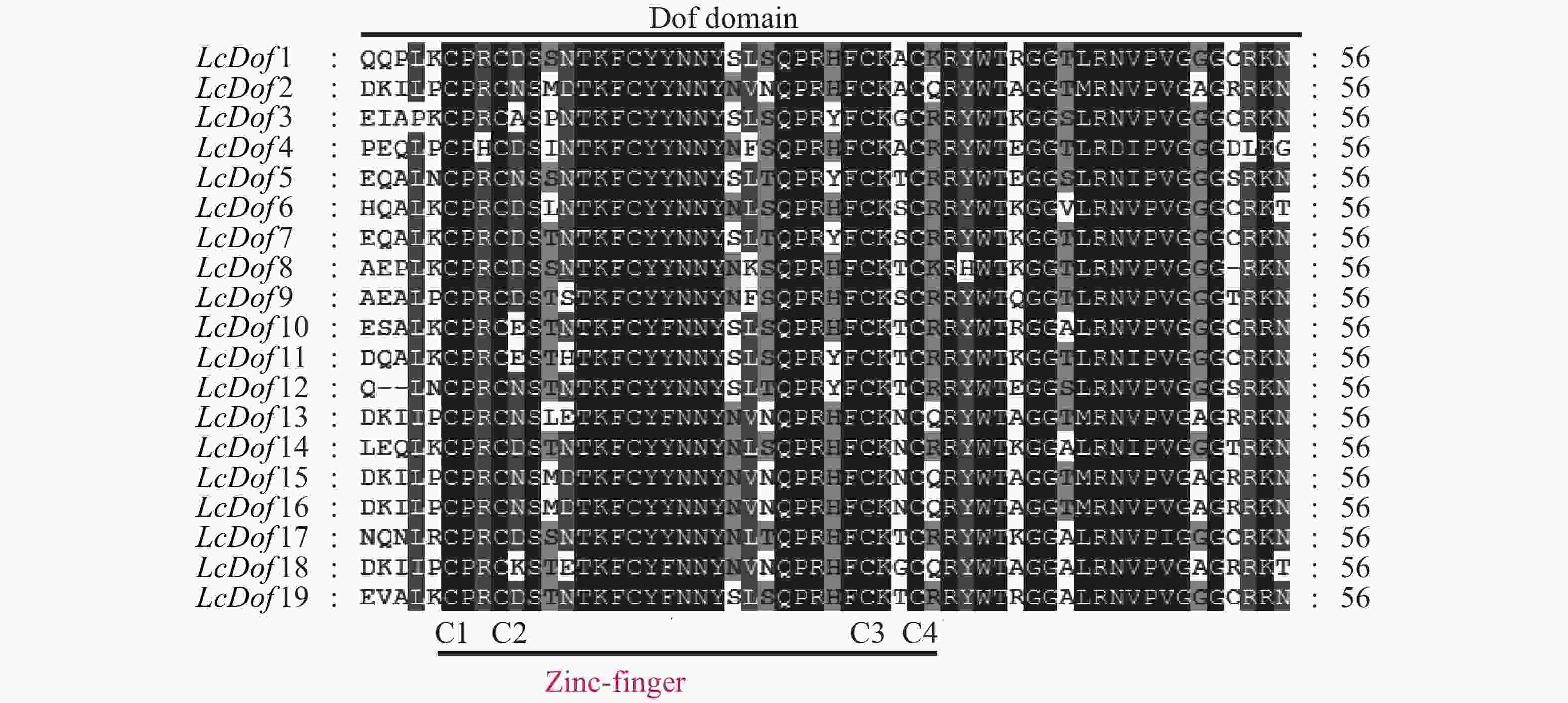
通过对荔枝Dof基因家族19个家族成员进行多重序列比对,抽取保守结构域进行观察分析(图2),发现19个LcDof蛋白结构域高度保守,均包含CX2CX21CX2C保守基序,构成了C2-C2型单锌指结构(Zinc-finger)。为进一步了解Dof基因家族在荔枝中的进化关系和生物学功能,分别以荔枝19个Dof(LcDof)、拟南芥中36个Dof(AtDof)和水稻中的30个Dof(OsDof)蛋白序列构建系统进化树 (图3)。聚类结果表明:19个荔枝Dof家族成员分别聚为4个亚家族(Group I-Group IV)个,其中GroupIV中含有LcDof基因家族成员数最多,有7个LcDof,分别为LcDof2、LcDof4、LcDof9、LcDof13、LcDof15、LcDof16、LcDof18,占基因家族总数的36.84%。其次为Group I,含有LcDof基因家族成员数最多,有6个LcDof,分别为LcDof1、LcDof3、LcDof7、LcDof11、LcDof12和LcDof15,占基因家族总数的31.58%。第三为Group II,含有5个LcDof基因家族成员,占基因家族总数的26.32%,分别为LcDof6、LcDof10、LcDof14、LcDof17、LcDof19。Group III含有LcDof基因家族成员最少,仅有1个LcDof8,占基因家族总数的5.26%。荔枝Dof家族成员与拟南芥Dof家族成员在进化上亲缘关系较近,而与水稻Dof家族成员关系较远,其中AtDof2.1和LcDof7,AtDof1.4和LcDof1,AtDof5.4和LcDof6,AtDof1.2和LcDof3为直系同源基因,推测以上荔枝中Dof与拟南芥的Dof在生物学功能上相似。LcDof5和LcDof12,LcDof10和LcDof19,LcDof4和LcDof9,LcDof2和LcDof15为旁系同源基因,推测荔枝Dof转录因子经历了基因复制事件,有可能存在功能的冗余。
-
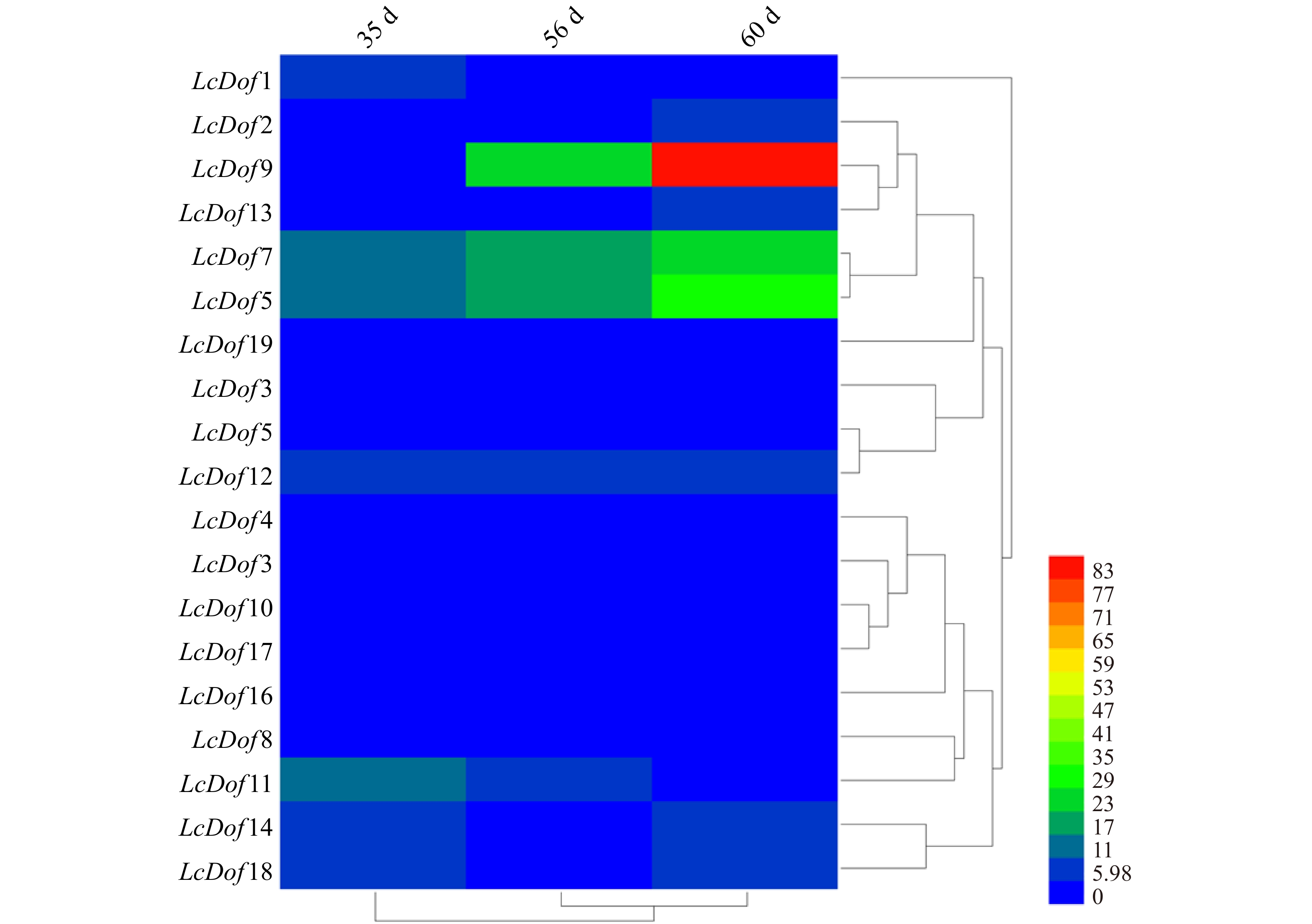
为研究荔枝Dof基因家族在荔枝果实不同发育时期的表达情况,利用妃子笑荔枝(果肉不同发育时期)的RNA-Seq 转录组数据库,找到候选的19个Dof基因对应转录本的RPKM值,然后用Heml热图软件对LcDofs的RPKM值取对数值转换制作聚类热图(图4)。图4显示,在果肉发育的不同时期均检测到19 个LcDof基因的表达,但表达丰度不同;其中LcDof7、LcDof9、LcDof12、LcDof15在果肉不同发育时期表达量均较强,而LcDof3、LcDof10、LcDof16、LcDof17、LcDof19表达量较低。Group Ⅰ中LcDof3、LcDof5、LcDof12表达规律相似,Group Ⅱ中LcDof6、LcDof10、LcDof17表达规律相似,Group Ⅳ中LcDo2、LcDof9、LcDof13和LcDof15表达规律相似,推测以上基因存在相近的基因功能。
2.1. LcDof的鉴定
2.2. LcDof蛋白的保守基序
2.3. Dof蛋白的多重序列比对及其进化关系
2.4. 荔枝Dof家族基因的表达
-
植物转录因子以多基因家族的形式存在,在植物生长发育及其基因表达调控过程中发挥重要的作用。随着高通量测序技术的发展,目前越来越多的物种基因组公布,在组学水平上对基因家族进行鉴定和生物信息学分析,已成为基因组学的研究热点。Dof转录因子作为植物中特有的一类转录因子,在植物生长发育和调控基因表达过程中发挥着重要的作用。目前关于植物Dof基因家族在包括模式植物拟南芥和水稻在内的多个物种中进行了鉴定和分析[5-19]。本研究参考拟南芥和水稻Dof家族基因的信息,基于荔枝果实发育转录组数据库,利用生物信息学方法,共鉴定了19个Dof基因家族成员。通过对LcDof基因家族中各蛋白的基本理化性质、保守基序、保守结构域、系统进化、基因表达进行了生物学分析。分析表明,LcDof转录因子编码的蛋白质为亲水性蛋白,定位于细胞核,这与马铃薯[11]、甜瓜[20]中的报道一致。通过系统进化分析,对荔枝Dof基因家族进行了详细的分类和命名。多重序列比对后利用Dof序列全长构建系统发育树,将LcDof分为4个不同的亚家族,该结果与在拟南芥、水稻、马铃薯、香蕉等物种上面的研究结果一致[5,11,15],拟南芥中Dof基因家族的37个成员、水稻的30个成员、马铃薯35个成员、香蕉中74个成员分为4个亚家族。进化树中聚类在1个亚家族的Dof含有的基序数目大致相似,比如亚家族4中的LcDof2、LcDof15和LcDof16,含有相同的基序,推测以上3个Dof具有相近的生物学功能。进化树中聚类在1个亚家族的Dof在荔枝果实发育中基因表达规律大致相似,例如亚家族1中LcDof3、LcDof5和LcDof12,推测以上3个Dof具有相近的基因功能。通过对荔枝Dof基因家族蛋白保守结构域分析可以看出,荔枝Dof蛋白的N−末端的DNA结合域具有高度的保守性,包含典型的保守结构域C2-C2的C2-C2型单锌指结构(Zinc-finger),且C−末端转录调控结构域复杂多变,这与前人的研究结果一致[5]。本研究同时对荔枝果实发育转录组中Dof蛋白的基因表达特征进行了全面的分析,其中进化关系近的LcDof成员表达规律相似,这与香蕉中的MaDof的研究结果类似[15]。








 DownLoad:
DownLoad: