-
橡胶树[Hevea brasiliensis (H. B. K.) Muell.-Arg.]原产于巴西亚马孙地区,19世纪引入亚洲热带地区[1],橡胶种植面积迅速扩大[2-4], 2017年世界橡胶种植面积达到1484万hm2[5]。我国橡胶人工林主要分布在海南和云南西双版纳,约占我国橡胶种植总面积的91.3%[6]。海南岛橡胶林的分布约占海南岛植被总面积的四分之一[7]。西双版纳是东南亚热带北缘重要的典型橡胶产区[8],西双版纳橡胶林约占西双版纳植被总面积的40%, 2010年西双版纳的橡胶人工林种植面积约50万hm2[9]。橡胶人工林已成为我国热带地区重要的森林生态系统,对维持热带陆地生态系统的平衡起着重要作用[10]。橡胶树受多种真菌致病因子的影响[11]。真菌根病以破坏橡胶林根系对土壤的水分和养分的吸收与利用方式对橡胶根系造成危害。此外,真菌根部疾病通常只有在感染扩散至根部死亡时才被观察到,此时多已超过有效治疗的程度[12]。
感染橡胶树根部的真菌病原体为白根病(Rigidoporus microporus)、褐根病(Phellinus noxius)和红根病(Ganoderma pseudoferreum)[13-14]。在经济上,白根病是橡胶树上最重要的病害,在全世界都造成橡胶产量的严重损失[15]。褐腐病和红腐病对橡胶林的经济影响较小,因为它们通常生长缓慢,对树木生长和乳胶生产的影响较小[13]。白根病可以感染整个热带地区的橡胶树,它是影响东南亚橡胶林的主要疾病之一[15-16]。健康树木接触到受感染的树根和木质碎屑,感染的根被大量的白色菌丝体覆盖,最初为褐色,后来为白色,因此,Rigidoporus microporus有白根病的俗称[16],在感染后期,真菌产生橙褐色子实体,新鲜时边缘为亮黄色,而树干底部周围的下表面变成红棕色[15]。在感染阶段,橡胶树的叶子开始变黄,最后这种疾病破坏了树冠和根系,导致树木死亡[16]。许多植物病原体感染橡胶树后无明显症状[17],并且可以在不利的环境条件下持续存在[18]。在疾病暴发之前,这些病原体通常基本上是潜在的。然而,这些病原体可以通过基于DNA的方法检测[19]。因此,基于DNA的方法可以全面了解潜在的植物病原菌群落。1978年,植物病理学家[20]将植物病原菌划分为:农杆菌属(Agrobacterium)、棒状杆菌属(Corynebacterium)、欧文氏菌属(Erwinia)、诺卡氏菌属(Nocardia)和假单胞菌属(Pseudomonas)等,归功于DNA测序技术和分类方法的进步,目前植物病原菌已有39个属[21]。
宏基因组测序技术较全面的反映了样本中微生物群落的组成。笔者通过宏基因组测序技术了解橡胶林土壤、根际、根表的病原微生物,并通过数据分析进一步了解微生物间的互作关系,对橡胶林土壤改良、提高橡胶树抗性和乳胶产量、并为橡胶林的病害预防及生长管理提供有效信息,对促进橡胶林生态多样性研究、病虫害的综合防控和橡胶产业的可持续发展具有理论和实践意义。
-
在海南(儋州、万宁和乐东)和西双版纳(景洪、勐仑和勐满)采集健康橡胶树的土壤和根系样本。
理化分析和土壤微生物采样:采样前用70%酒精取样器具需进行消毒灭菌处理。去除表面浮土,采样员戴上无菌手套,以橡胶树树干为圆心(半径为0.5 m),在圆上随机取3个点,去除表层5 cm的土壤,在5~20 cm土层采集橡胶树根际周围土壤200 g,去除可见杂质,用2 mm筛网过筛处理,然后将3份土壤混合均匀,分别用自封袋装500 g土壤作为理化分析样品和取5~10 g土壤装入无菌离心管中,一并放入保温盒中带回实验室,所有样品置于干冰寄送至美吉生物公司测序。
根际微生物取样:采样前用70%酒精对取样器具进行消毒灭菌处理。用消毒的剪刀在土壤微生物采样的3个点上分别剪取5~8根直径2 mm以内细根系,每根根系长约9~12 cm(尽量不要碰伤表皮),每个点在根系周围1 mm采集橡胶树根际土壤,把根际土壤抖落在无菌自封袋中,去除植物根、动物残骸及其他杂质,过0.85 mm筛,将3份土样混合均匀,取5~10 g装入无菌离心管,一并低温运至实验室,所有样品置于干冰中,送美吉生物公司测序。
根表微生物取样:采样员戴上无菌手套,在根际微生物取样中剪取的根系置于保温盒内保存,24 h内送回实验室。在实验室用无菌刷轻轻刷去根系表层附着的土壤,将根系置于无菌离心管中,浸没于无菌PBS缓冲溶液,在180 r·min−1下清洗20 min;用无菌镊子取出根系组织,将根系再次置于无菌离心管中加入无菌PBS缓冲溶液,在180 r·min−1下清洗20 min;取出根系组织,将根系再次置于无菌离心管中加入无菌PBS缓冲溶液,超声波洗涤10 min(超声波清洗器设置:160 W,30 s/30 s,10 min)。将3次洗涤液置于无菌离心管中,用 12 000 g离心10 min,收集沉淀(根表微生物)并置于−80 ℃冰箱保存。所有根表微生物样品置于干冰中至美吉生物公司测序。
-
土壤理化和养分测定参考鲁如坤[22]的方法。土壤理化性质测定了土壤含水率和pH。土壤养分测定主要有土壤有机质、速效氮(氨态氮和硝态氮)、速效磷、速效钾、全氮、全磷、全钾(表1)。土壤有机碳测定采用重铬酸钾-外加热法;全氮测定采用靛酚蓝比色法;全磷测定采用钼锑抗比色法;全钾采用火焰光度计法;速效钾采用醋酸铵提取火焰光度计法;速效磷测定采用盐酸氟化铵法。硝态氮测定采用氯化钠提取法,氨态氮测定采用靛酚蓝比色法。
环境因子 S_01 S_02 S_03 S_04 S_05 S_06 速效磷(AP)/(mg·kg−1) 21.16 1.95 1.36 5.02 3.47 2.64 有机质(SOM)/(g·kg−1) 7.87 15.38 10.22 35.20 22.01 16.64 硝态氮(NO3N)/(mg·kg−1) 13.82 15.34 4.27 18.37 7.30 7.44 氨态氮(NH4N)/(mg·kg−1) 6.33 14.46 18.96 22.11 25.41 15.16 速效钾(AK)/(mg·kg−1) 20.07 28.72 54.47 35.06 78.50 39.31 pH 3.76 4.28 4.44 3.78 4.90 4.72 含水量(WC)/% 14.48 19.37 19.82 29.77 25.19 16.68 全磷(TP)/(g·kg−1) 0.55 0.86 0.49 1.38 1.07 0.67 全氮(TN)/(g·kg−1) 0.72 1.48 1.03 2.67 1.78 1.22 全钾(TK)/(g·kg−1) 12.96 11.23 33.94 8.34 17.83 6.91 Table 1. Nutrients of soil samples by metagenome sequencing
-
采用 Illumina Hiseq PE150/250 高通量测序平台。使用fastp软件对序列进行质控,得到clean reads序列,然后利用Megahit软件进行序列组装,得到的组装序列通过MetaGene软件进行基因预测(序列长度≥100 bp),利用CD-HIT软件和SOAPaligner 软件进行聚类(90% identity、90% coverage)和基因的丰度计算(95% identity),比对NR数据库获得样本的物种分类学注释信息,通过 NR 库中的分类学信息数据库获得物种注释结果,计算物种的丰度,并统计各个样品在界、门、纲、目、科、属和种的分类学水平上物种的丰度,从而构建相应分类学水平上的丰度表。比对病原与宿主互作数据库(PHI),对橡胶林土壤、根际及根表进行植物病原物种注释,得到橡胶林土壤、根际及根表植物病原物种信息,并进行生物信息学分析。
-
RDA 即冗余分析,是约束化的 PCA 分析,从图中可以直观地看出样本分布和环境因子间的关系。RDA主要用于分析物种或功能与环境因子之间关系,RDA 是基于线性模型。使用R语言vegan包中RDA分析和作图。主坐标分析(PCoA),是一种非约束性的数据降维分析方法,研究样本群落组成的相似性或差异性, PCoA是基于所选距离矩阵进行作图。本研究选用bray_curtis距离算法,通过降维的方式来找出影响样本群落组成差异的潜在主成分。使用R语言PCoA分析作图。
-
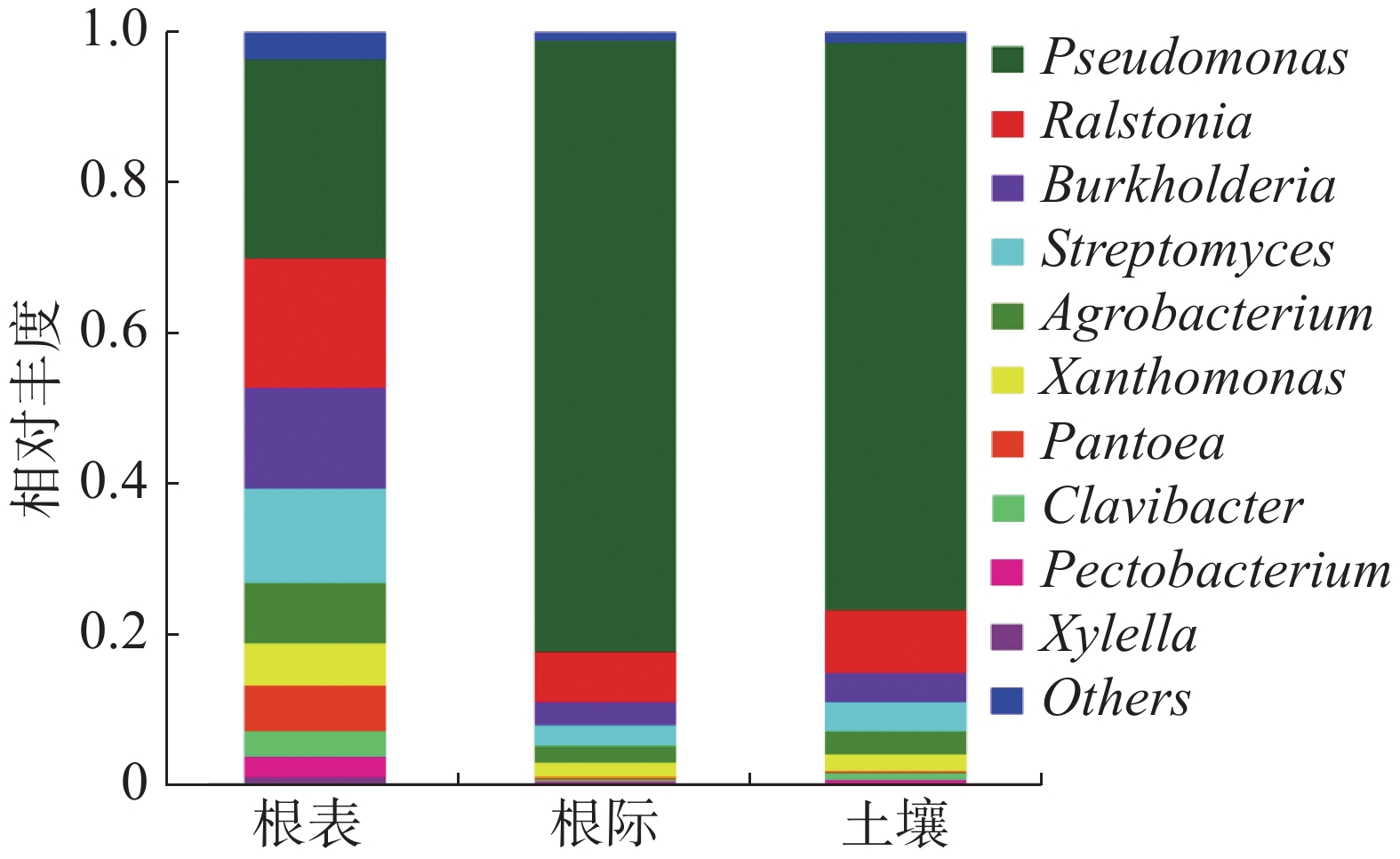
通过宏基因组测序结果与病原和宿主互作数据库比对注释,得到植物病原微生物在土壤、根际及根表的组成,病原微生物在门、属和种水平分别为:4、30和53(表2)。由图1可知,植物病原微生物在门的水平上属于:变形菌门(Proteobacteria)、放线菌门(Actinobacteria)和子囊菌门(Ascomycota),其中变形菌门在土壤、根际及根表的平均占比为90.67%,根际样本占比最大,根表样本占比最少;放线菌门在土壤、根际及根表的平均占比为7.67%,根表样本占比最多,根际样本占比最少;子囊菌门在土壤、根际及根表的平均占比为1.11%,根表样本占比最多,根际样本占比最少。
门 属 种 S_01 S_02 S_03 S_04 S_05 S_06 R_01 R_02 R_03 R_04 R_05 R_06 RP_01 RP_02 RP_03 RP_04 RP_05 RP_06 Actinobacteria Clavibacter Clavibacter_
michiganensis136 168 258 112 46 54 128 150 52 102 36 74 484 476 1034 120 36 74 Actinobacteria Streptomyces Streptomyces_
scabiei974 682 912 792 258 246 496 732 418 1148 264 382 1278 1616 1134 732 426 516 Ascomycota Zymoseptoria Zymoseptoria_
tritici12 10 8 4 2 0 2 10 6 8 6 2 12 10 4 8 12 2 Ascomycota Leptosphaeria Leptosphaeria_
maculans0 0 0 0 0 2 0 0 0 0 4 0 4 0 0 2 10 0 Ascomycota Parastagonospora Parastagonospora_nodorum 12 4 0 0 0 0 0 2 2 14 0 0 2 4 0 6 0 4 Ascomycota Alternaria Alternaria_
alternata0 4 8 0 0 2 0 4 10 0 0 0 0 0 4 0 0 0 Ascomycota Bipolaris Bipolaris_
maydis0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 4 0 Ascomycota Bipolaris Bipolaris_
sorokiniana6 0 6 12 0 0 0 4 0 24 0 0 0 0 2 10 0 4 Ascomycota Bipolaris Bipolaris_
zeicola0 0 0 0 0 0 0 0 0 0 0 0 4 0 0 0 0 2 Ascomycota Pyrenophora Pyrenophora_
tritici-repentis10 4 6 0 0 0 2 2 6 0 2 0 0 0 0 0 0 0 Ascomycota Setosphaeria Setosphaeria_turcica 0 0 0 6 2 2 0 0 0 8 0 0 6 2 0 0 8 4 Ascomycota Penicillium Penicillium_
expansum0 0 0 14 0 0 2 0 0 6 0 2 0 0 0 10 0 0 Ascomycota Botrytis Botrytis_cinerea 0 12 6 6 0 6 0 18 8 8 0 0 4 0 0 2 0 4 Ascomycota Sclerotinia Sclerotinia_
sclerotiorum22 8 2 6 0 0 0 4 30 2 2 16 4 8 4 12 6 2 Ascomycota Colletotrichum Colletotrichum_
gloeosporioides6 6 0 10 4 10 4 6 0 6 2 4 12 8 0 16 2 2 Ascomycota Colletotrichum Colletotrichum_
graminicola0 6 0 4 0 0 0 12 0 8 0 4 0 6 0 4 0 0 Ascomycota Colletotrichum Colletotrichum_
higginsianum8 0 0 2 2 0 0 0 2 2 0 0 2 0 0 0 0 0 Ascomycota Colletotrichum Colletotrichum_
orbiculare0 4 6 0 0 4 0 6 0 6 0 0 30 42 12 14 8 8 Ascomycota Claviceps Claviceps_
purpurea10 0 0 0 0 0 0 8 0 0 0 0 14 0 0 0 0 0 Ascomycota Metarhizium Metarhizium_
robertsii22 14 0 2 0 10 12 18 4 2 8 2 2 6 6 2 8 0 Ascomycota Fusarium Fusarium_fujikuroi 0 2 0 0 0 0 0 6 0 0 0 2 0 0 0 4 0 0 Ascomycota Fusarium Fusarium_
oxysporum86 38 10 28 0 0 12 60 8 20 4 8 4 22 12 10 0 4 Ascomycota Fusarium Fusarium_
verticillioides0 0 2 16 2 0 0 0 0 0 2 0 2 0 2 8 0 0 Ascomycota Fusarium Nectria_haematococca 12 0 4 0 2 0 2 10 4 4 0 0 2 0 12 4 4 0 Ascomycota Gaeumannomyces Gaeumannomyces_graminis 50 12 2 24 6 4 14 14 6 2 4 2 2 8 60 8 2 2 Ascomycota Grosmannia Grosmannia_clavigera 10 0 2 0 0 2 6 2 0 2 0 0 0 4 8 4 2 14 Ascomycota Rosellinia Rosellinia_
necatrix10 26 4 30 2 10 26 22 10 54 24 6 28 56 40 36 2 20 Basidiomycota Puccinia Puccinia_
striiformis24 0 16 0 2 2 0 0 0 10 0 0 0 0 0 0 0 22 Proteobacteria Agrobacterium Agrobacterium_
tumefaciens280 518 242 462 164 224 244 598 234 518 196 226 310 478 332 380 236 218 Proteobacteria Agrobacterium Agrobacterium_
vitis86 146 112 216 64 82 104 188 132 222 56 106 146 146 146 200 78 154 Proteobacteria Burkholderia Burkholderia_
cenocepacia480 738 610 590 298 340 790 760 438 604 242 352 1008 2858 208 566 362 414 Proteobacteria Burkholderia Burkholderia_
glumae140 126 104 110 118 94 196 104 96 134 108 162 74 158 50 130 76 90 Proteobacteria Ralstonia Ralstonia_
solanacearum1460 2024 1450 1294 1060 852 1274 1794 1480 1304 1046 1032 1050 1710 614 1402 888 804 Proteobacteria Dickeya Dickeya_
dadantii14 40 14 28 2 4 10 52 18 38 8 14 24 22 24 10 8 26 Proteobacteria Dickeya Dickeya_
solani20 30 12 24 24 8 0 14 22 40 30 14 136 6 6 20 10 24 Proteobacteria Erwinia Erwinia_
amylovora4 14 6 6 2 4 4 10 2 18 0 2 128 60 122 6 14 4 Proteobacteria Pantoea Pantoea_ananatis 12 2 10 10 12 2 2 2 0 18 2 2 558 398 4294 2 10 42 Proteobacteria Pantoea Pantoea_
stewartii56 32 18 20 34 36 56 22 48 50 28 22 152 80 576 18 40 64 Proteobacteria Pectobacterium Pectobacterium_
atrosepticum18 6 0 0 2 0 4 0 0 0 0 0 4 12 10 2 4 4 Proteobacteria Pectobacterium Pectobacterium_
carotovorum68 96 74 86 34 42 58 106 54 62 20 24 194 142 494 62 60 202 Proteobacteria Pectobacterium Pectobacterium_
wasabiae4 12 0 16 4 0 0 14 4 12 0 2 28 46 34 6 8 6 Proteobacteria Pseudomonas Pseudomonas_
aeruginosa800 1070 636 686 884 974 1304 990 1318 772 766 1032 540 702 568 552 458 420 Proteobacteria Pseudomonas Pseudomonas_
cichorii20 4 6 30 8 2 16 20 10 32 2 6 16 16 12 10 8 6 Proteobacteria Pseudomonas Pseudomonas_
fluorescens696 804 668 758 13374 20058 18386 982 24716 834 10646 18296 942 946 728 692 794 558 Proteobacteria Pseudomonas Pseudomonas_savastanoi 0 10 10 2 10 24 14 18 28 0 20 10 2 4 0 6 4 4 Table 2. Pathogenic microorganisms in soil, rhizosphere and rhizoplane of rubber plantations in Hainan and Xishuangbanna

Figure 1. Relative abundance of pathogenic microorganisms in soil, rhizosphere and rhizoplane at the phylum level
由图2可知,病原微生物在属水平上属于:假单孢菌属(Pseudomonas)、劳尔氏菌属(Ralstonia)、伯克霍尔德氏菌属(Burkholderia)、链霉菌属(Streptomyces)、农杆菌属(Agrobacterium)、黄单胞菌属(Xanthomonas)、泛菌属(Pantoea)、麦角菌属(Clavibacter)、果胶杆菌属(Pectobacterium)和木质部小菌属(Xylella)。假单孢菌属在土壤、根际及根表的占比为61.20%,根际样本占比最大,根表样本占比最少;劳尔氏菌属在土壤、根际及根表的占比为10.70%,根际样本占比最少,根表样本占比最大;伯克霍尔德氏菌属在土壤、根际及根表的占比为6.80%,根际样本占比最少,根表样本占比最大;链霉菌属在土壤、根际及根表的占比为6.38%,根际样本占比最少,根表样本占比最大;农杆菌属在土壤、根际及根表的占比为4.45%,根际样本占比最少,根表样本占比最大。根表样本中病原菌占比相对平均,而根际样本中病原菌主要为假单孢菌属,占比达到81.44%。在属水平上主要以子囊菌门为主。
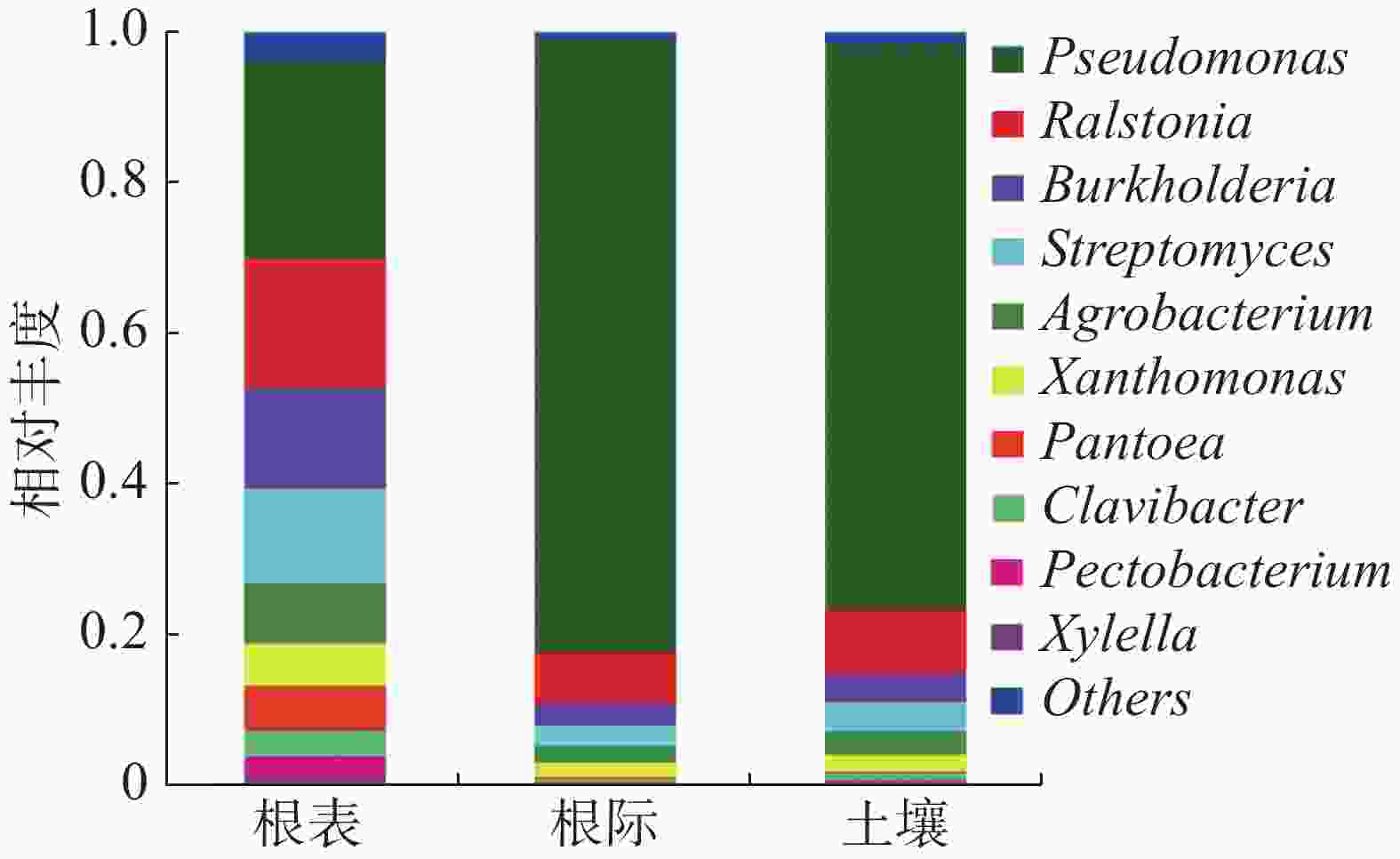
Figure 2. Relative abundance of pathogenic microorganisms in soil, rhizosphere and rhizoplane at the genus level
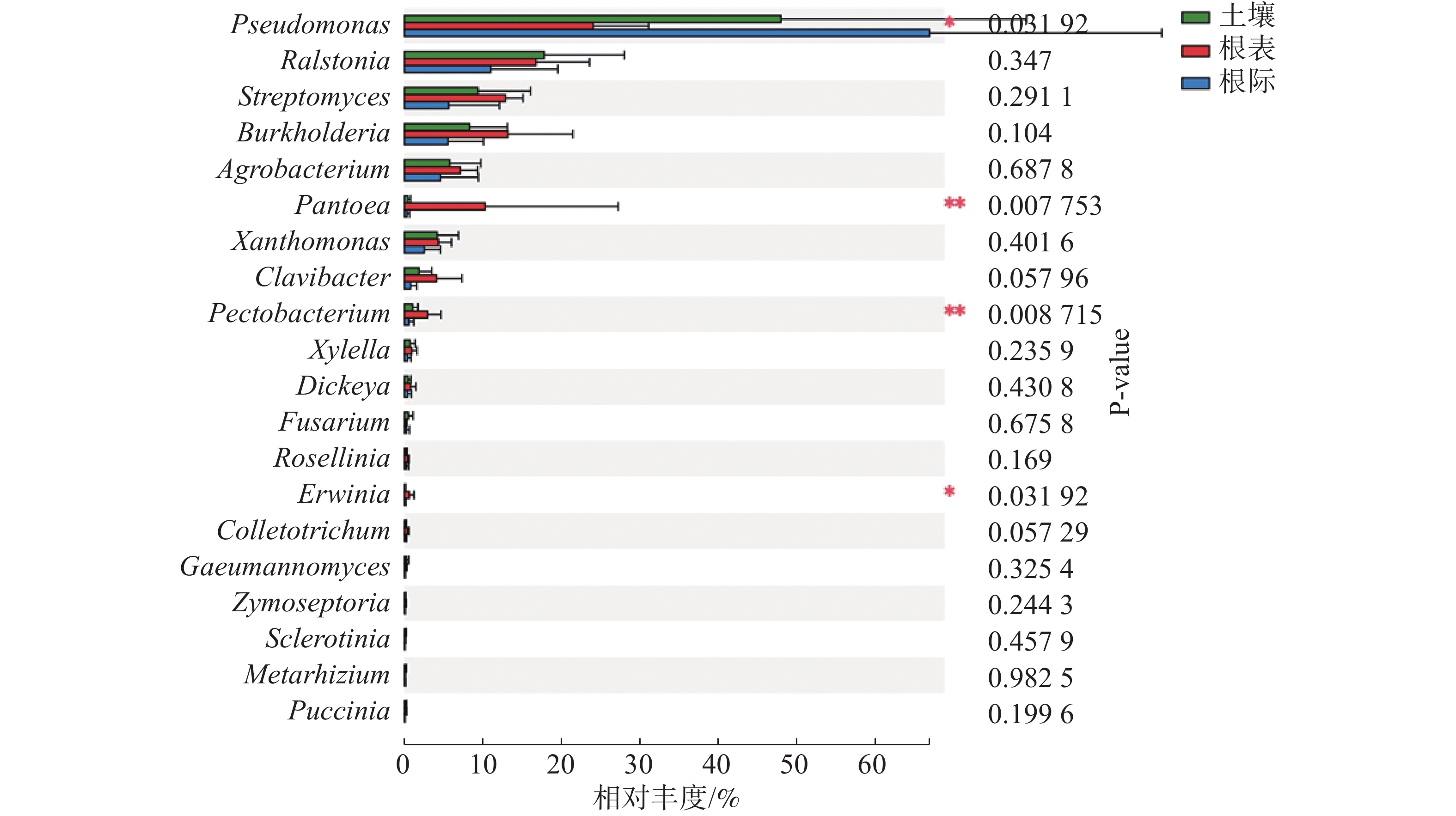
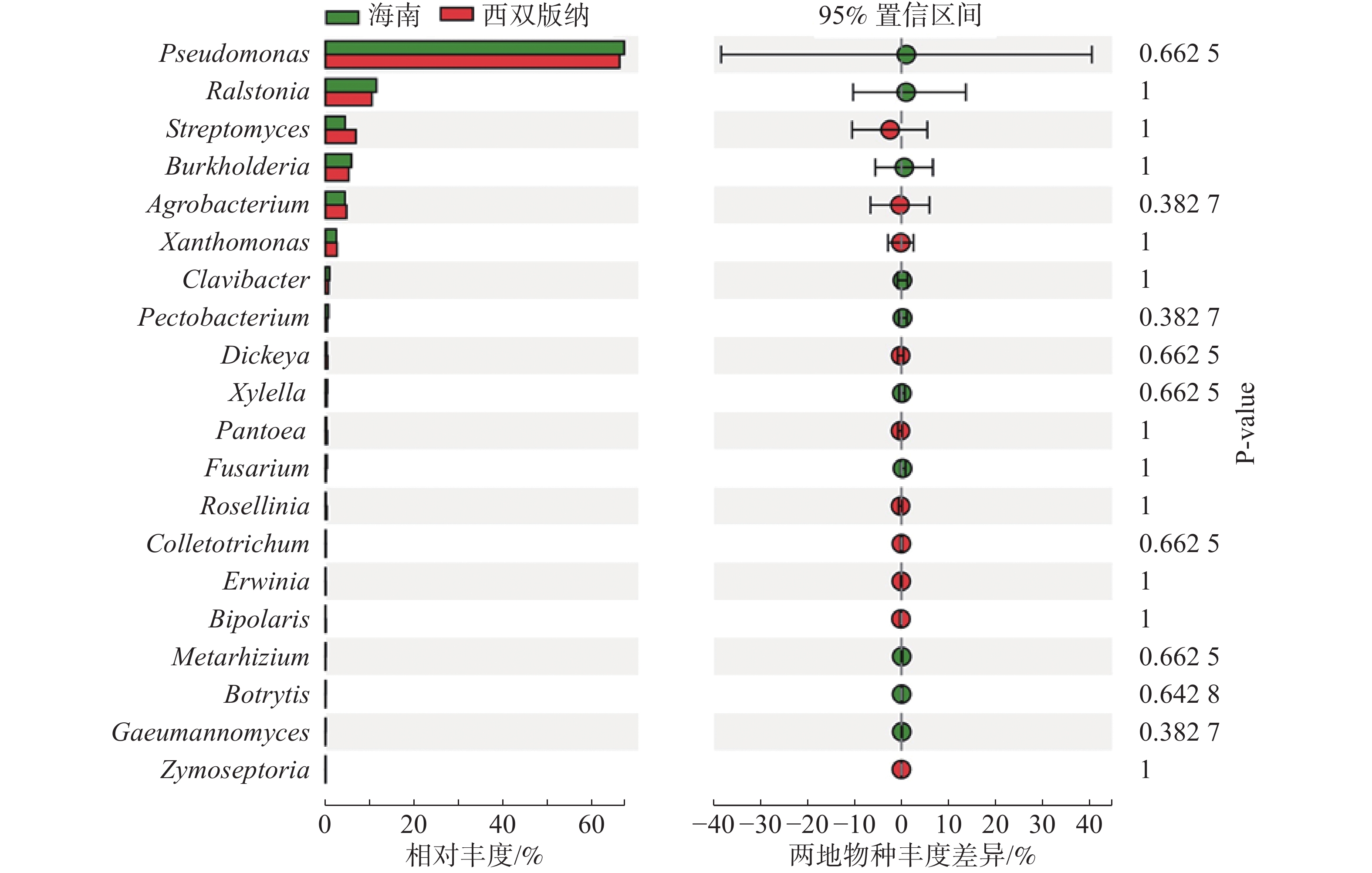
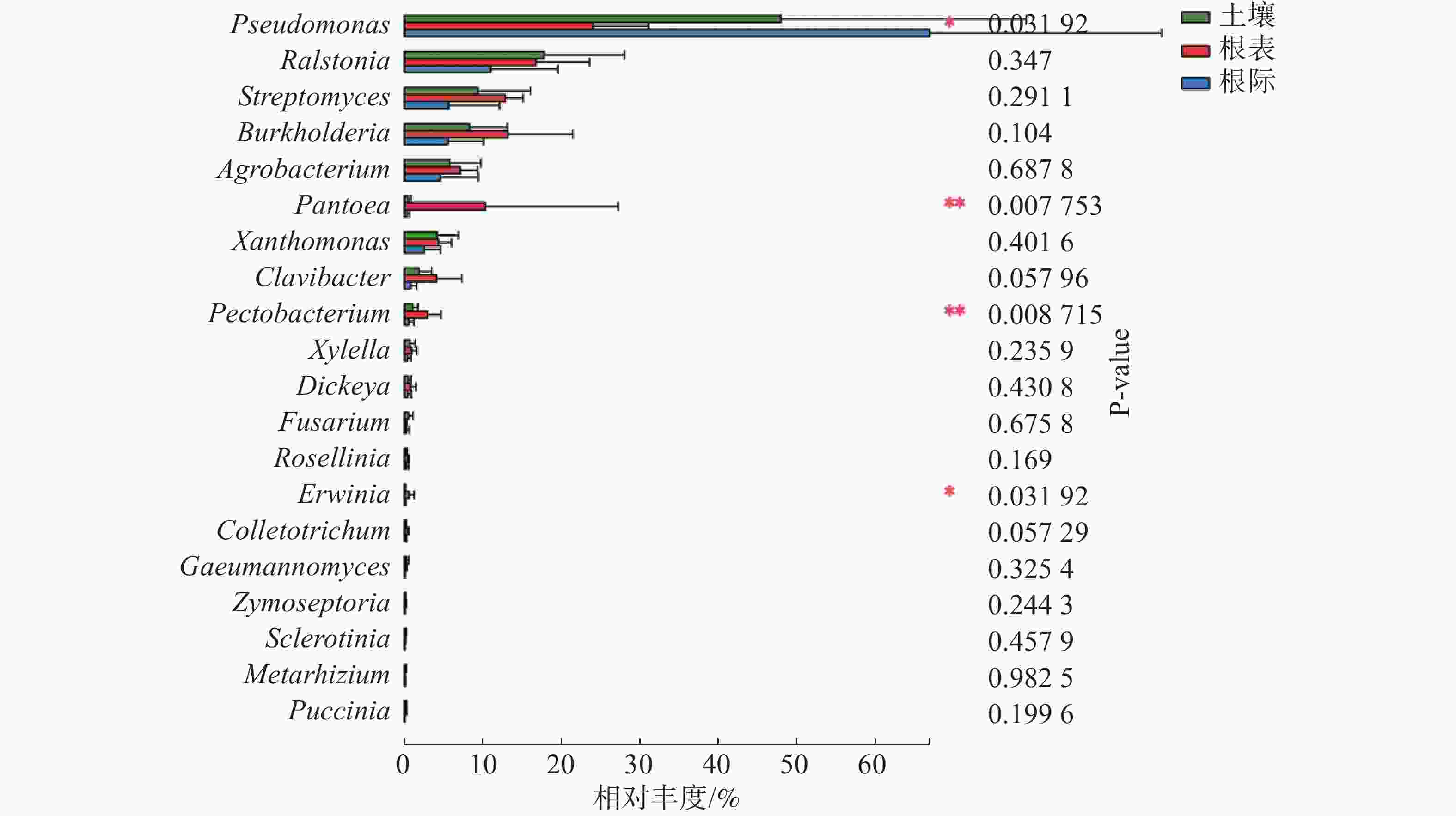
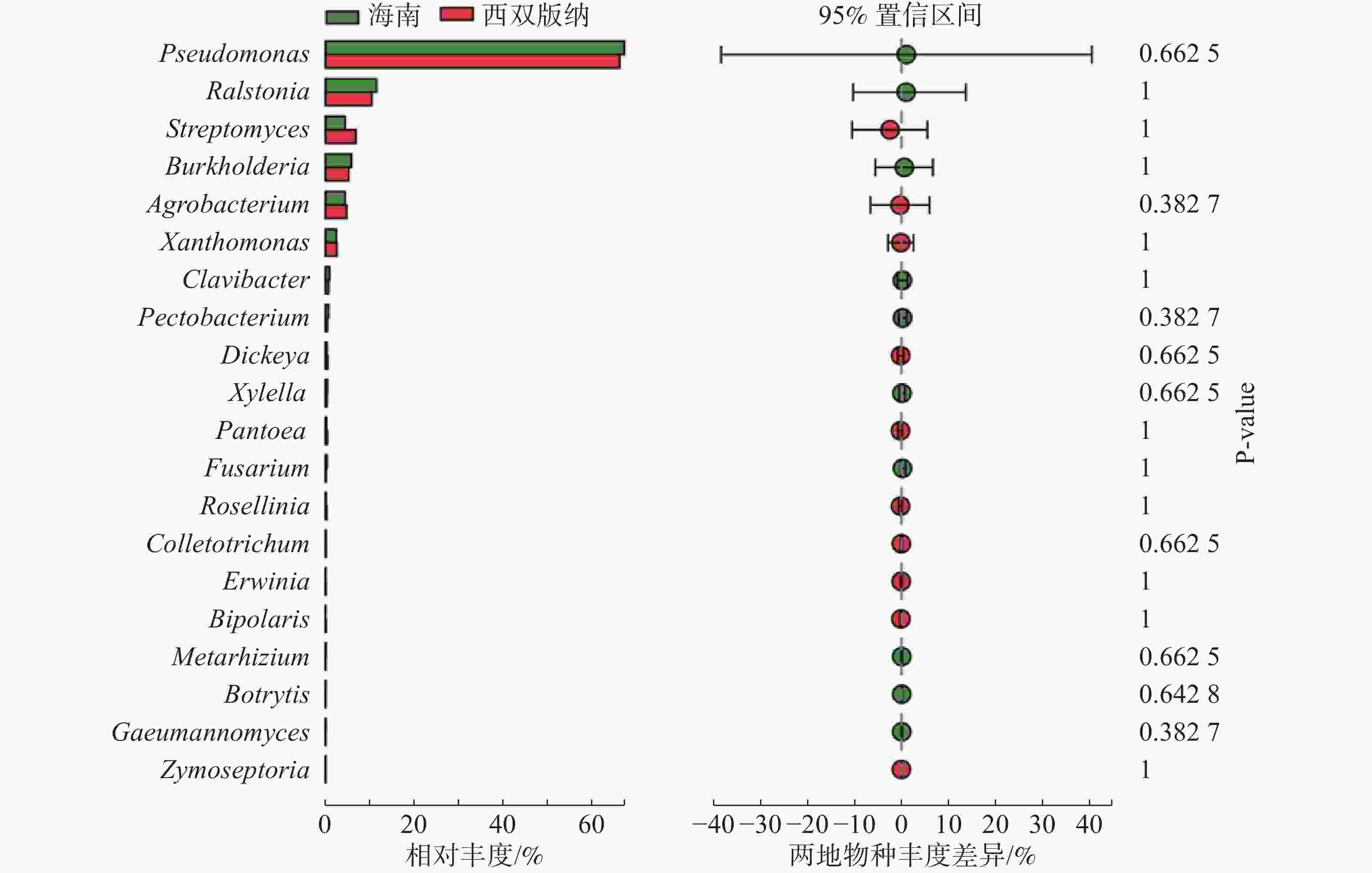
由图3可知,在土壤、根际及根表样本属水平上,假单孢菌属和欧文氏菌属(Erwinia)有显著差异(P=0.03),泛菌属和果胶杆菌属有极显著差异(P<0.01)。其中假单孢菌属在根表样本中的丰度明显小于土壤和根际样本,欧文氏菌属、泛菌属和果胶杆菌在根表样本中的丰度明显大于土壤和根际样本。由图4可知,两地根际病原微生物在属水平上无显著差异。
-
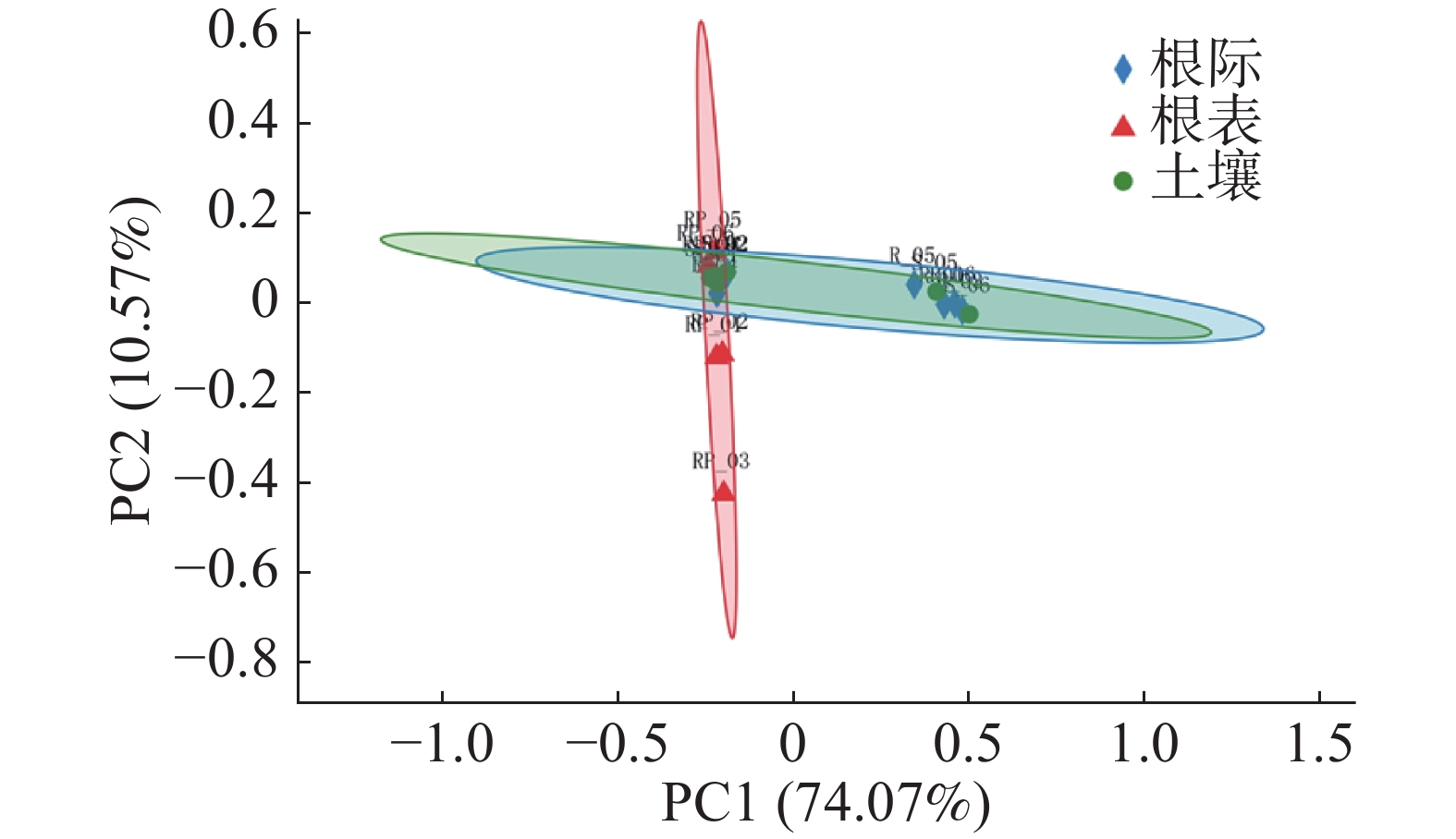
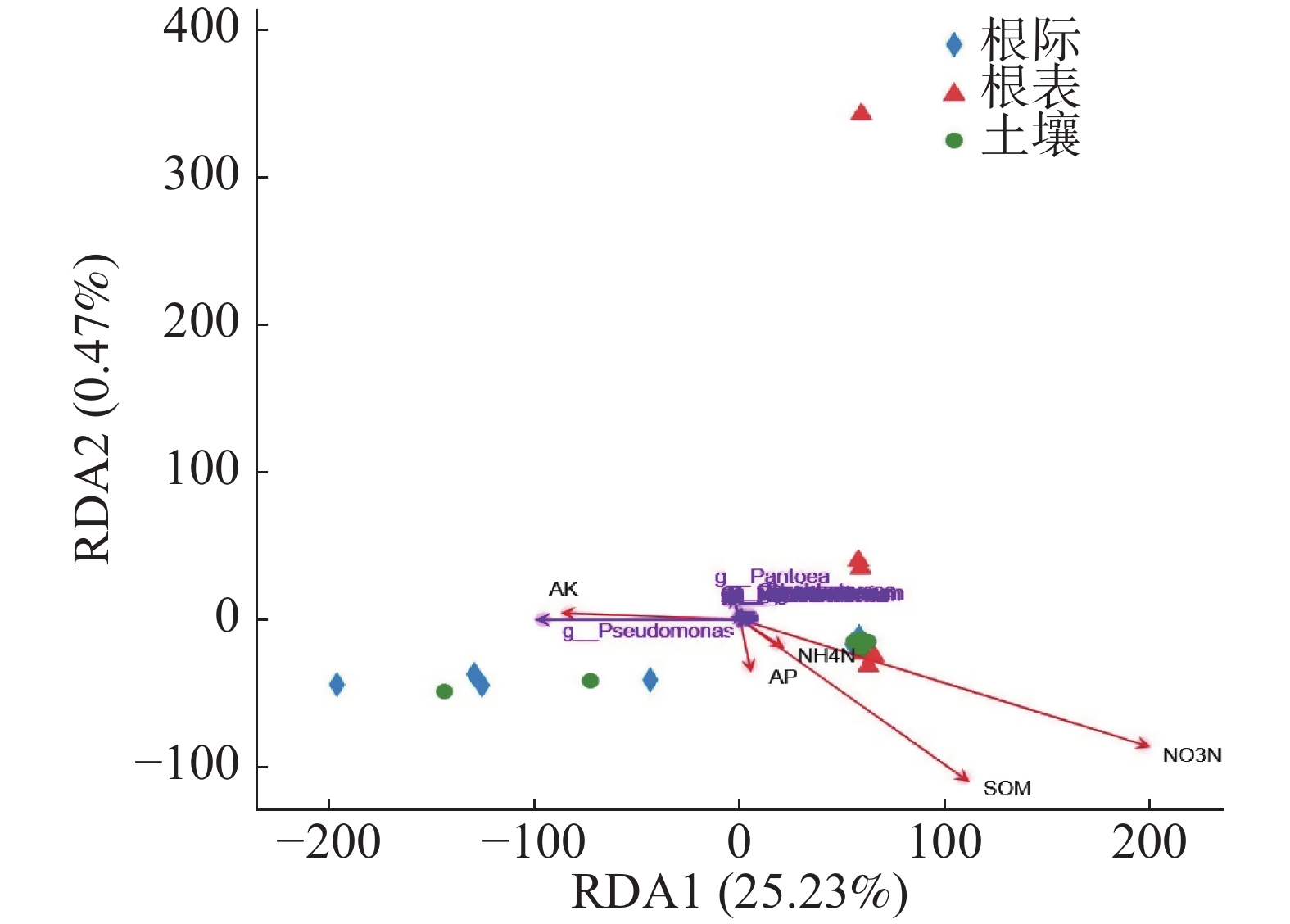
由图5可知,PC1轴解释74.07%,PC2轴解释10.57%。由图5和图6可知,根表样本离散程度低,说明病原微生物在根表样本中分布均匀,而土壤和根际样本则相反。土壤和根际样本置信椭圆高度重合,说明土壤和根际样本病原微生物差异更小,根表样本在PC1轴高度聚类,也说明病原微生物在根表样本中高度相似,但乐东的根表样本有一点差异。
-
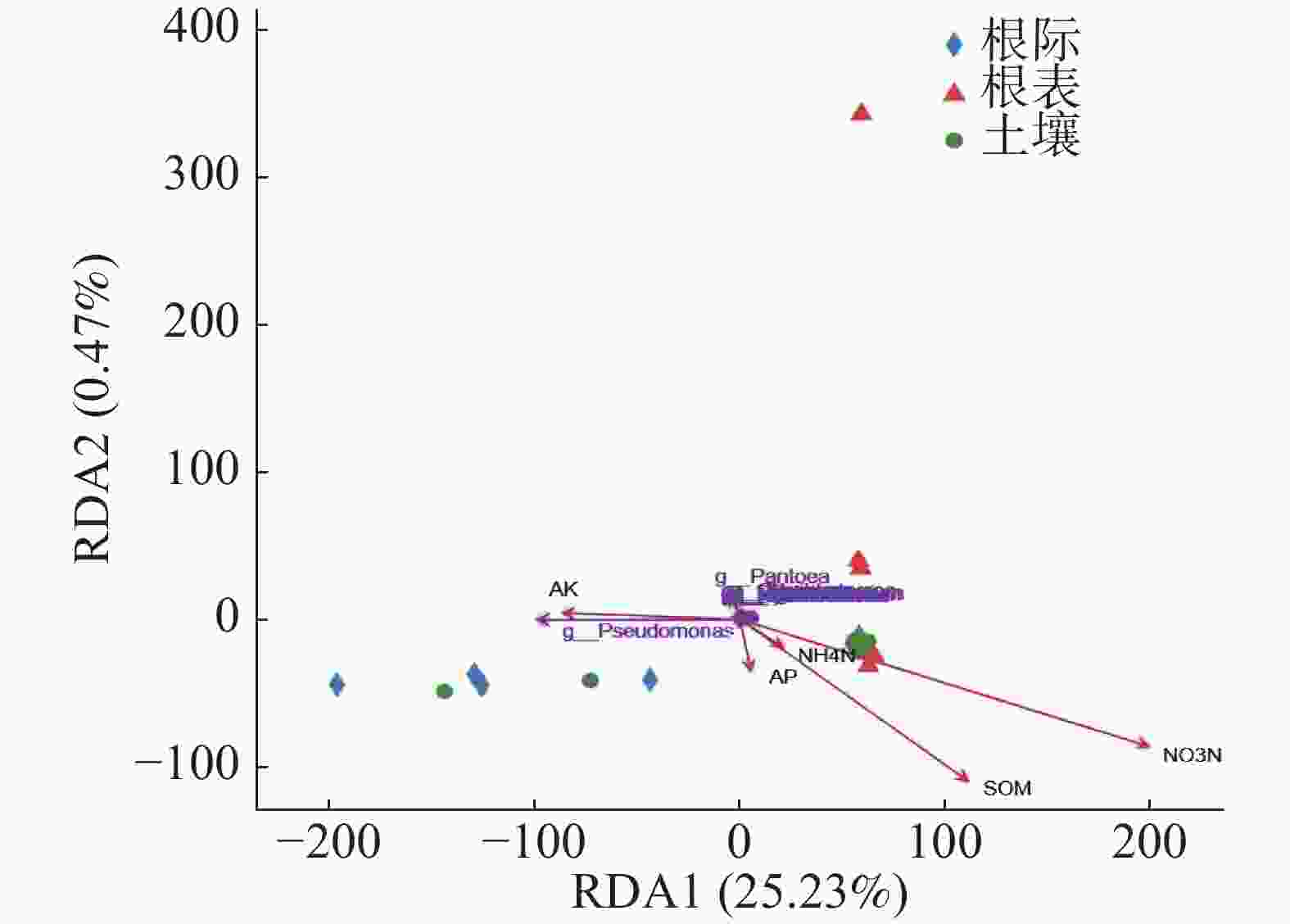
基于病原微生物属水平RDA分析,由图6可知,RDA1轴解释25.23%,RDA2轴解释0.47%,勐仑和勐满土壤和根际样本病原微生物组成相似,乐东根表样本与其他根表样本有差异。由图6和表3可知,只有硝态氮与病原微生物呈显著正相关(P<0.05),其他环境因子与病原微生物无显著关系。
RDA1 RDA2 R2 P_values AP 0.41 −0.91 0.01 0.944 SOM 0.70 −0.71 0.17 0.302 NO3N 0.82 −0.57 0.30 0.039 NH4N 0.71 −0.70 0.00 0.97 AK −0.93 0.36 0.04 0.761 pH −0.99 0.13 0.10 0.522 WC 0.85 −0.53 0.10 0.507 TP 0.72 −0.69 0.25 0.128 TN 0.77 −0.64 0.18 0.277 TK −0.47 0.88 0.28 0.099 Longitude −0.03 1.00 0.12 0.361 Latitude 0.10 −0.99 0.16 0.241 Elevation 0.07 −1.00 0.11 0.395 Table 3. Correlation factors for RDA analysis of pathogenic microorganism
-
通过宏基因组测序结果与病原和宿主互作数据库比对注释,得到植物病原微生物在土壤、根际及根表的组成。研究表明[11],橡胶树受到多种真菌致病因子的影响,而本实验结果表明,健康土壤中,土壤、根际及根表样本中均未出现引起橡胶树根病的真菌,说明健康土壤中有抑制致病菌的有益微生物,这与Weller等[23]研究结果相似。有研究表明[24],多年生作物不断地从土壤中吸收养分,导致土壤肥力下降,而土壤养分缺乏会增加植物对感染的敏感性,本实验结果表明,硝态氮与病原微生物呈显著正相关,其他环境因子与病原微生物无显著关系。那么在对橡胶林施肥时,是否可以增加氨态氮的比例,从而减少病原微生物的种类和丰度。
实验结果表明,植物病原微生物在门的水平上属于变形菌门、放线菌门和子囊菌门,其中变形菌门在根表样本占比最少,而放线菌门和子囊菌门在根表样本占比则最多,表明放线菌门和子囊菌门的病原微生物更适应根表环境,或者说根表环境吸引放线菌门和子囊菌门的病原微生物定殖,表明不仅有益微生物会被植物根系招募,某些病原微生物也会被植物根系招募。土壤和根际样本病原微生物组成相似,各根表样本病原微生物组成相似,表明病原微生物的组成可能与根表环境有关。在属水平上,病原微生物以子囊菌门种类最多,表明子囊菌门的病原微生物在橡胶树根系中分布广泛。假单孢菌属主要分布在土壤和根际,欧文氏菌属、泛菌属和果胶杆菌主要分布在根表。两地根际病原微生物在属水平上无显著差异。
Analysis of pathogenic microorganisms in rubber plantations based on macrogenome sequencing
doi: 10.15886/j.cnki.rdswxb.2022.01.005
- Received Date: 2021-05-25
- Accepted Date: 2021-11-11
- Rev Recd Date: 2021-11-05
- Available Online: 2022-01-13
- Publish Date: 2022-01-31
-
Key words:
- rubber plantation /
- metagenome sequencing /
- soil /
- root /
- pathogenic microorganisms
Abstract: In order to understand the composition and influencing factors of broad-spectrum pathogenic microorganisms in soil and root (rhizosphere and rhizoplane) of rubber plantations, metagenome sequencing was used to analyze the composition and influencing factors of pathogenic microorganisms in soil, rhizosphere and rhizoplane of healthy rubber plantations in Hainan (Danzhou, Wanning and Ledong) and Xishuangbanna (Jinghong, Menglun and Mengman). The results showed that plant pathogenic microorganisms belonged to Proteobacteria, Actinobacteria and Ascomycota at the phylum level, and to Pseudomonas, Ralstonia, Burkholderia, Streptomyces, Agrobacterium, Xanthomonas, Pantoea, Clavibacter, Pectobacterium and Xylella at the genus level with Ascomycota as the main pathogen at the genus level. There was no significant difference in pathogenic microorganisms in the rhizospheres between the rubber plantations in Hainan and Xishuangbanna. The composition of pathogenic microorganisms was similar in the soil and rhizosphere samples, and similar in the rhizoplane samples. There was a significant positive correlation between NH4-N and pathogenic microorganisms. In conclusion there was no significant difference in pathogenic microorganisms in soil, rhizosphere and rhizoplane between the rubber plantations in Hainan and Xishuangbanna. A significant positive correlation was observed between pathogenic microorganisms and NH4-N, and that the composition of pathogenic microorganisms was related to the exudates of the rhizoplane.
| Citation: | QUAN Fei, LAN Guoyu, WEI Yaqing, LI Mingmei, SUN Shuqing, DU Haonan. Analysis of pathogenic microorganisms in rubber plantations based on macrogenome sequencing[J]. Journal of Tropical Biology, 2022, 13(1): 27-35. doi: 10.15886/j.cnki.rdswxb.2022.01.005 |








 DownLoad:
DownLoad: