-
芋螺(Conus)是分布于热带海洋中的肉食性软体动物,全世界约有800种[1]。不同种类的芋螺都能通过分泌各自特有的肽类毒素(芋螺毒素、芋螺肽)捕食鱼类、虫类或其他软体动物。根据芋螺的食性,分为食鱼芋螺,食虫芋螺和食螺芋螺3大类[2]。芋螺的毒液是由长而卷曲的毒管上皮细胞分泌产生的。当芋螺捕食猎物时,毒腺的肌肉蠕动,推动毒液进入鱼叉状的舌状牙齿中,将毒液快速注射入猎物体内[3-6]。研究发现,每种芋螺毒液中含有约3 000种不同的芋螺毒素肽。在萼托芋螺(Conus episcopatus)的1个个体毒液中,竟发现了3 305个前体毒素基因序列[7]。由此预估全球芋螺可产生超过150万种的天然毒素肽,它们具有调节各种离子通道和受体等的特殊功能[8],已成为神经科学研究工具药和治疗新药研发的重要来源[9]。然而迄今为止,发现的芋螺毒素还不足其总量的1%,因而大量的新型芋螺毒素尚有待进一步开发。目前,获取芋螺毒素序列的方法主要有4种,包括粗毒液纯化[10]、芋螺毒素基因克隆[11]、毒管转录组测序[7, 12]、毒液蛋白质组学和多组学联用测序分析[13-14]。多组学是一种通过生物信息学将转录组学与蛋白质组学整合的综合策略[15],在芋螺毒素研究领域很受欢迎。芋螺毒素的研究需要持续不断地捕捞活体芋螺,但由于气候变暖,芋螺赖以生存的海洋环境也越来越恶化,再加上芋螺壳工艺品国际贸易的兴起,造成了芋螺资源的严重破坏,70%以上的芋螺遭遇生存危机。所以人们更应重视芋螺资源的保护和可持续利用,例如可建立不同芋螺种类的基因组DNA文库、毒管cDNA文库以及毒素肽库,长期保存并加以利用。粗毒纯化需要大量的芋螺,且只能发现一些表达量较高的毒素,为克服粗毒纯化的局限性,20世纪90年代,基因克隆为新型芋螺毒素的发现提供了极大的帮助[11]。由于芋螺毒素是由特定的前体基因表达的,所以可以用特定的引物通过PCR技术进行扩增[16-20]。因而,简单快速地获取高质量的芋螺基因组DNA是关键。有关海南产信号芋螺基因组DNA的提取方法优化的研究目前尚未见报道,为此,笔者拟从信号芋螺(Conus litteratus)的不同组织器官中,选取4种试剂盒,分别提取其基因组DNA,然后比较各种提法的效果,并对其中的优选试剂盒的提取条件进行优化,旨在建立简便快速提取信号芋螺基因组DNA的方法。
-
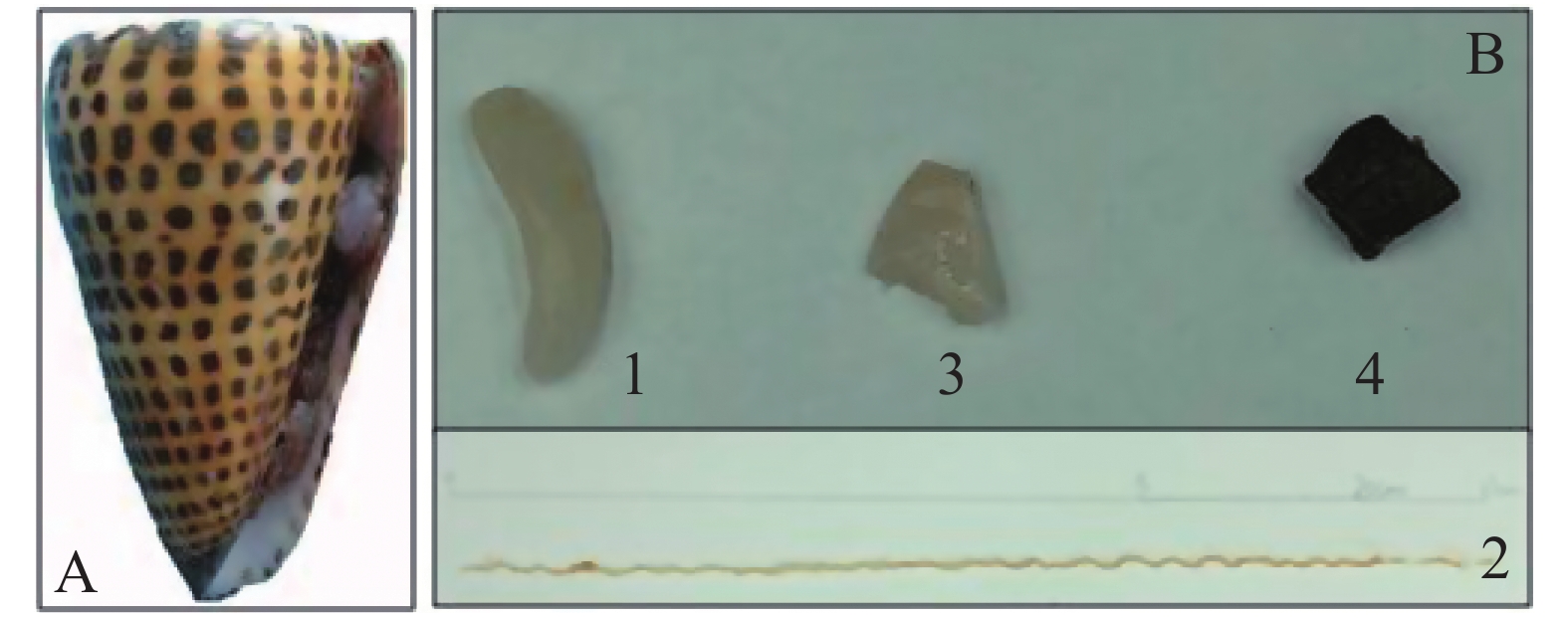
信号芋螺(Conus litteratus)活体采自中国南海海域(N15°46′~17°08′,E111°11′~112°54′),于−80 ℃超低温冰箱保存(图1A),将冷冻的信号芋螺置于冰上30 min,待微解冻后破壳,解剖,分别取出毒腺、毒管、腹足以及肝胰脏(图1B)4种组织待用。
磁珠法动物基因组DNA抽提试剂盒(B518721)和改良的DNA快速提取试剂盒(B518221)均购于上海生工公司;FastPure Tissue DNA提取试剂盒(DC102)购于诺唯赞公司;海洋动物组织DNA提取试剂盒(DP324)购于天根公司。2×Taq PCR Master Mix(天根生化科技有限公司);4S Red Plus核酸染色剂(生工生物工程有限公司);琼脂糖、6×Loading Buffer, 10×Loading Buffer(北京索莱宝科技有限公司);DL15000 DNAMarker,DL5000 DNAMarker(南京诺唯赞公司)。
-
所用仪器(品牌)主要有:琼脂糖水平电泳仪(Bio-Rad),高速冷冻离心机(HITACHI),PCR扩增仪(Eppendorf Mastercycler X50),低温循环水浴锅(Polyscience),凝胶成像系统(Protein-Simple),NanoDrop 2000超微量分光光度计(Thermo)等。
-
称取毒腺20 mg、毒管10 mg、腹足20 mg及肝胰脏10 mg,冰上剪碎,分别置于研钵中用液氮研磨成粉末,加入1.5 mL的离心管中,然后向离心管中加入200 μL Buffer MCL,400 μL Buffer MACL和20 μL Proteinase K(PK),上下颠倒混匀,稍离心,65 ℃水浴30~40 min。期间,多次颠倒混匀,待溶液基本澄清,取出离心管,12 000 r·min−1离心5~10 min,小心吸取上清液500 μL至新的1.5 mL的离心管中;加入20 μL RNase (10 g·L −1)溶液,混匀,室温放置5 min。加入500 μL Buffer MA和15 μL MagicMag Beads(MMB),MMB加到液面以下,且尽量将枪头上残留的MMB吸干净,混匀,室温静置1 min。将离心管置于磁力架上30 s,待MMB完全吸壁后,吸弃上清,取下离心管;加700 μL 70%乙醇,颠倒混匀,置于磁力架上30 s,吸弃上清,重复1次。室温开盖干燥或者55 ℃恒温箱开盖干燥至管内无液体;向离心管中加入50 μL TE Buffer(pH8.0),65 ℃水浴5~10 min,混匀;取出离心管置于磁力架上30 s,小心吸取上清至新的离心管,即得基因组DNA。
-
组织处理如1.3,加入400 μL buffer Digestion和20 μL PK,振荡混匀;65 ℃水浴1 h至溶液变澄清透明,否则可适当延长裂解时间;水浴后加入20 μL RNase(10 g·L−1)溶液,颠倒混匀,室温放置5 min;加入200 μLBuffer PA,充分颠倒混匀,置于20 ℃冰箱5 min后室温10 000 r·min−1离心5 min,吸取500 μL上清液到新的离心管中(若上清液浑浊,可加等体积的氯仿混匀,12 000 r·min−1离心取上清);加入等体积的异丙醇,充分颠倒混匀,于−20 ℃放置20 min,10 000 r·min−1离心5 min,弃上清;用1 mL 75%乙醇漂洗2 min,10 000 r·min−1离心2 min,取上清,重复1次,漂洗时一定要使沉淀悬浮起来;开盖室温倒置至残留乙醇完全挥发,肉眼可见DNA沉淀变成半透明状,离心管内无乙醇味道即可;干燥完成后用50 μL TE Buffer(pH8.0)溶解即得DNA。
-
组织处理如1.3,加入230 μL Buffer GA和20 μL PK,涡旋混匀,55 ℃水浴至组织完全酶解;加入20 μL RNase (10 g·L−1)溶液,颠倒混匀,室温放置5 min;若消化液比较浑浊或存在明显的颗粒,则10 000 r·min−1离心3 min,转移上清液至新的离心管中;加入250 μl Buffer GB,充分涡旋混匀,70 ℃水浴10 min;加入250 μL无水乙醇,涡旋混匀;将gDNA Columns吸附柱置于收集管中;将上步所得的混合液转移到吸附柱中,10 000 r·min−1离心1 min;弃滤液,用500 μL Washing Buffer A洗涤1次,10 000 r·min−1离心1 min,650 μL Washing Buffer B洗涤2次,弃滤液,空管离心2 min;将吸附柱置于新的1.5 mL离心管中,加50 μL预热至70 ℃的Elution Buffer至吸附柱的膜中央,室温放置3 min,10 000 r·min−1离心1 min;弃掉吸附柱,离心管中即为DNA。
-
组织处理如1.3,加入200 μL GA涡旋振荡15 s;加入20 μL RNase (10 g·L−1)混匀,放置5 min。加入20 μL PK,涡旋混匀,简短离心以去除管盖内壁的水珠;56 ℃水浴30 min,待溶液变澄清透明后加入200 μL缓冲液GB,充分颠倒混匀,会产生一些白色沉淀,70 ℃放置10 min,溶液变清亮;加入200 μL无水乙醇,充分颠倒混匀,此时可能出现絮状沉淀;将吸附柱CB3放入收集管中,将上步所得溶液和沉淀都加入吸附柱中,12 000 r·min−1离心30 s,弃废液,将吸附柱放回收集管中;加入500 μL缓冲液GD,12 000 r·min−1离心30 s,弃废液;再用600 μL漂洗液PW漂洗2次,12 000 r·min−1离心30 s,弃废液,将吸附柱放入收集管中,空管离心2 min;将吸附柱置于室温数分钟,彻底晾干吸附柱中残留的漂洗液;将吸附柱放入新的1.5 mL的离心管中,向吸附柱中央悬空滴加50 μL 洗脱缓冲液TE,室温放置5 min,12 000 r·min−1离心2 min,离心管中所得即为DNA。
-
取DNA溶液1 μL,通过NanoDrop 2000紫外分光光度计检测DNA的浓度和吸光度,并根据组织用量,计算DNA的产率。再取9 μL DNA溶液,加1 μL的10×Loading Buffer用于凝胶电泳检测,TAE缓冲液,制1%的胶,电压90 V,电泳时间30 min,用于观察样品的相对分子质量大小。
-
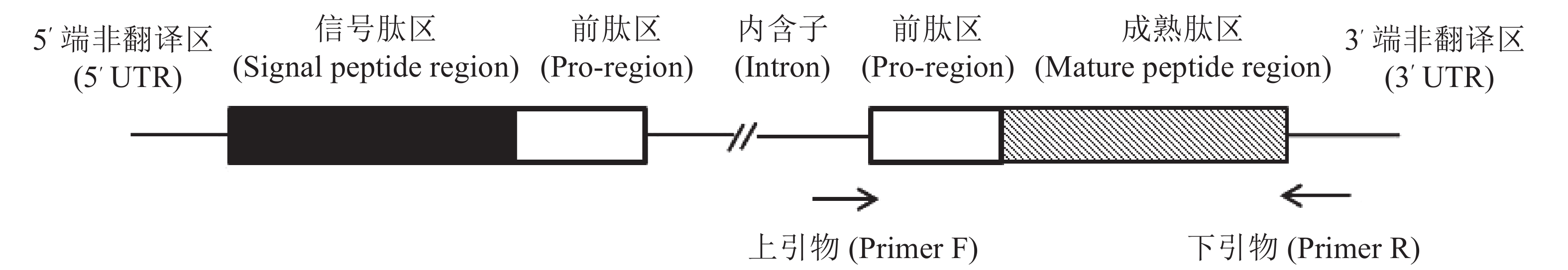
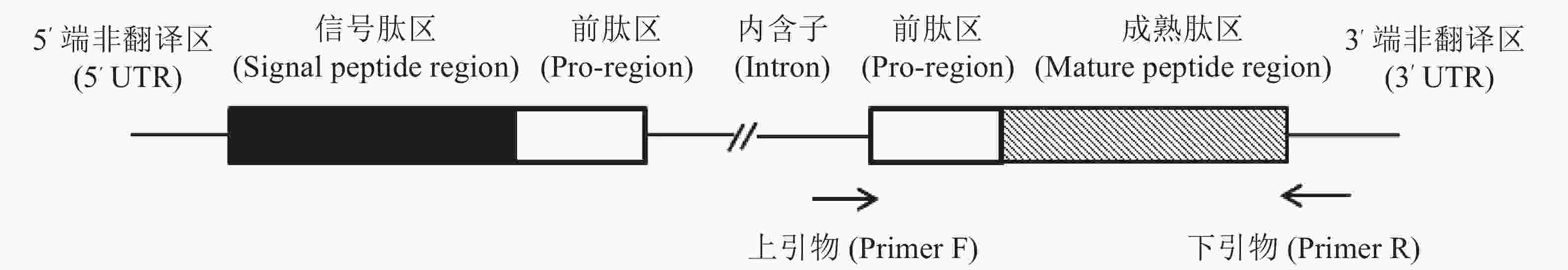
以芋螺核基因DNA上α−芋螺毒素基因设计引物,F: 5′-GTGGTTCTGGGTCCAGCA-3′, R: 5′-GTCGTGGTTCAGAGGGTC-3′;正向引物根据内含子的保守序列设计,反向引物则是根据3′-UTR设计(图2),4种方法所提的DNA为模板,判断所提DNA是否为核基因组DNA,该PCR反应体系为:25 μL: 2×Taq PCR Master Mix 12.5 μL,引物各0.5 μL,DNA模板1 μL,补加ddH2O至25 μL。反应条件:预变性,95 ℃,5 min;循环内:变性,95 ℃,30 s;退火,56 ℃,30 s;延伸,72 ℃,1 min,共30个循环;再延伸72 ℃,10 min。将所得PCR产物进行1%琼脂糖凝胶电泳检测,电压90 V,电泳时间30 min。
-
从表1可知,磁珠法DNA抽提试剂盒所提取的毒腺、毒管、腹足以及肝胰脏的A260/A280分别为1.69,2.01,1.73,1.61,其中,毒管和肝胰脏组织提取的纯度较低;改良的DNA快速提取试剂盒法的A260/A280分别为1.89,1.91,1.84,1.69,提取效果比磁珠法好,但肝胰脏有降解和蛋白污染等的问题;FastPure Tissue DNA提取试剂盒法的A260/A280分别为1.67,1.94,1.84,1.87,毒腺的A260/A280值偏低;海洋动物组织DNA提取试剂盒法的A260/A280分别为1.80,1.82,1.74,1.63,肝胰脏有明显的蛋白污染。比较4种方法的A260/A280可以看出,改良的DNA快速提取试剂盒提取的DNA纯度和海洋动物组织DNA提取试剂盒方法整体略优于其他2种方法,而且不同芋螺组织中,毒腺和腹足提取DNA的纯度更高。
提取方法
Method样品名
Sample质量/mg
QualityA260/A280 浓度/(mg·L−1)
Concentration产率/(mg·kg−1)
Yield磁珠法试剂盒 毒腺 19 1.69 16.3±1.7 42.9±4.5 毒管 19 2.01 247.7±28.5 651.8±75 腹足 24 1.73 58.1±1.5 121.0±3.1 肝胰脏 11 1.61 350.2±9.3 1 590.9±42.3 改良DNA快速提取试剂盒 毒腺 25 1.89 68.9±10.2 137.8±20.4 毒管 12 1.91 109.9±21.1 457.9±87.9 腹足 20 1.84 80.3±9.8 200.8±24.5 肝胰脏 11 1.69 388.3±19.4 1 765±88.1 FastPure DNA提取试剂盒 毒腺 23 1.67 39.5±13.9 85.9±30.2 毒管 21 1.94 179.2±30.8 426.7±73.3 腹足 22 1.84 37.3±10.2 84.7±23.2 肝胰脏 12 1.87 357.7±20.7 1 490.4±86.3 海洋动物组织DNA提取试剂盒 毒腺 20 1.80 27.3±5.3 68.3±13.3 毒管 20 1.82 195.4±12.5 488.5±31.25 腹足 25 1.74 71.2±9.4 142.4±18.8 肝胰脏 10 1.63 162.1±28.9 810.5±144.5 注:质量和吸光度为均为3次测量的均值。
Note: Weight and A260/A280 are a mean of three measured values.Table 1. The purity and yield of genome DNA of Conus litteratus extracted from the tissues of different organs
-
从表1可知,改良的DNA快速提取试剂盒法整体优于其他试剂盒法,其中,毒管和肝胰脏组织的产率明显高于毒腺和腹足,腹足则略高于毒腺,结合A260/A280数值,4种方法提取腹足的基因组DNA纯度最高,A260/A280数值均接近1.80,因此,改良的DNA快速提取试剂盒法提取腹足组织能够获得较高纯度且较高产率的DNA(图3)。
-
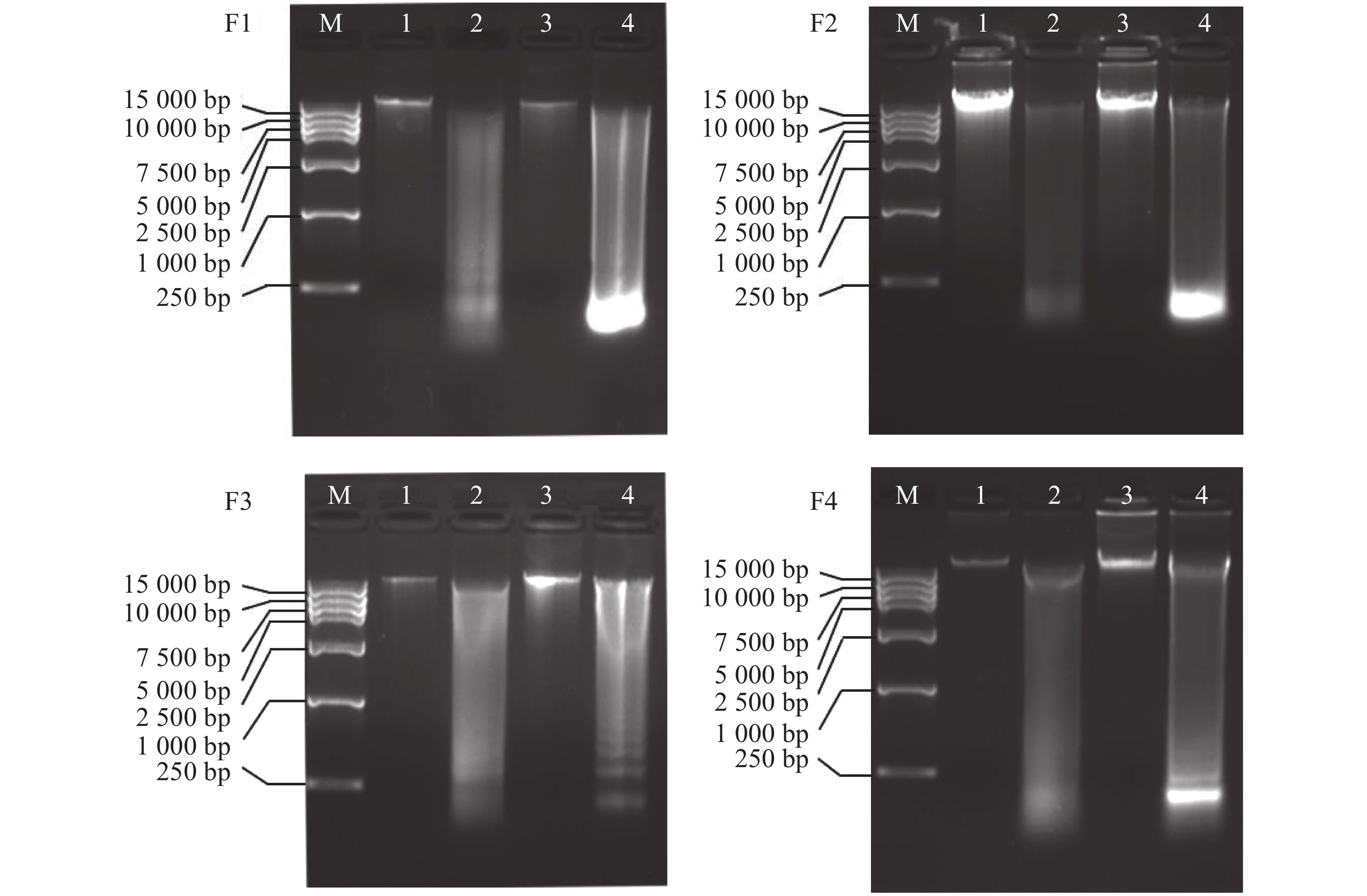
将4种方法提取的不同组织的基因组DNA进行水平电泳检测(图4),从电泳条带看,条带均比较清晰。其中,改良的DNA快速提取试剂盒法电泳图中,毒腺,腹足提取的DNA条带明亮且清晰,大小均超过15 kb,DNA完整性较好且不存在明显污染情况;而毒管和肝胰脏DNA则完整性较差,条带弥散,且肝胰脏泳道最下方有明亮的条带,应是RNA污染,此结果在提取肝胰脏中较为常见,毒管泳道最下方也有模糊条带,可见毒管也有RNA污染的问题,毒腺和腹足组织DNA无明显的污染条带,此结果和NanoDrop 2000检测结果相符。这可能是由于毒腺和腹足密度较大,组织无法研磨充分,阻碍了蛋白质溶解,故而裂解过程比较困难,影响了DNA的产率。肝胰脏最易研磨,故而产率最高,但是由于其组织过于柔软,且富含DNA酶,在冷冻、解剖、以及提取过程中可能造成其DNA降解,故而电泳条带多呈现弥散状。
Figure 4. Agarose gel electrophoresis of genomic DNA of Conus litteratus different tissues extracted with four different kits
通过4种试剂盒的电泳图(图4)可以发现,毒管和肝胰脏的DNA的降解严重,纯度低,对比毒腺与腹足的DNA产率,腹足略优于毒腺,况且毒腺组织材料相对少,不适用于大量提取DNA,所以腹足是更为合适的提取组织,且从图中(图3)可发现,提取腹足DNA时,改良的DNA快速试剂盒提取的DNA产率最高,因此,改良的DNA快速提取盒法提取腹足的DNA产率高、完整性好、纯度高。提取信号芋螺的腹足基因组DNA用该方法更合适。
-
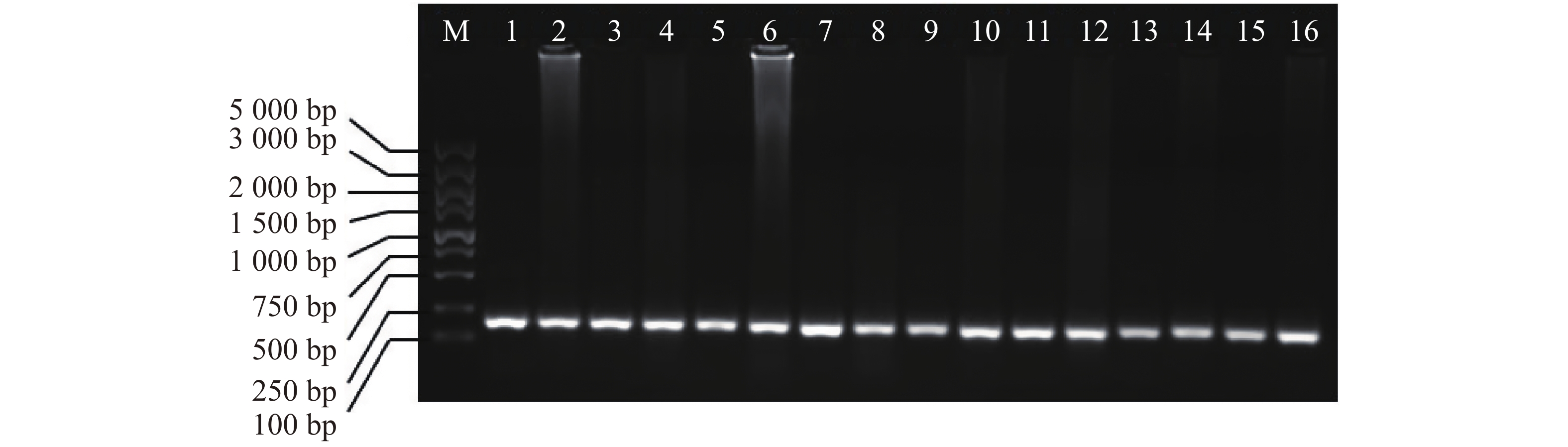
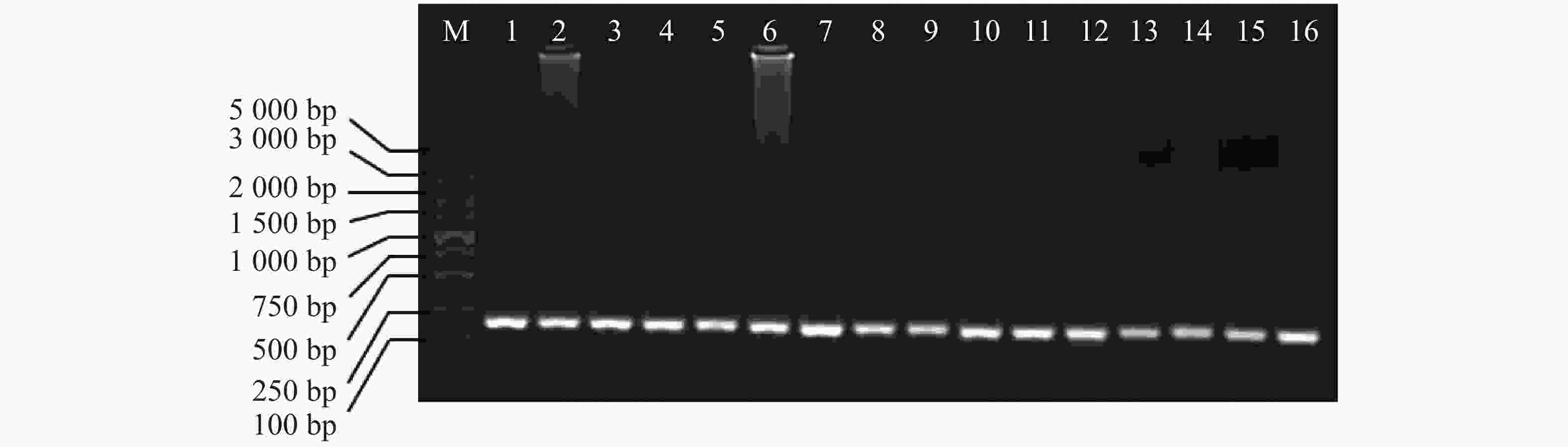
以芋螺核基因特有的A超家族的α−芋螺毒素基因为引物,4种不同的方法提取的DNA为模板进行PCR,电泳发现1~16泳道均有明亮的条带,大小在100~250 bp之间(图5),与芋螺毒素的前体基因相符,测序后确定目的条带为α−芋螺毒素基因,大小为167~170 bp。因此,所提取的DNA可用于后续实验。
-
DNA提取试剂盒种类繁多,应用方法多样,其中,包括吸附材料结合法(材料包括磁珠、硅质材料和阴离子交换树脂等)、浓盐法、阴离子去污剂法以及苯酚抽提法等。试剂盒提取DNA的操作过程大多比较简单,一般用时约1 h,组织用量为1~100 mg,其主要步骤包括匀浆,裂解细胞释放DNA,去除蛋白和其它杂质,纯化和DNA洗脱[21]。不同组织处理方式和提取的效果也不同,其中,芋螺的毒腺,腹足因富含胶原蛋白需要用液氮研磨,肝胰脏则因为富含DNA酶也需要用液氮充分研磨至粉末,并且尽量减少暴露在空气中的时间以减少降解。组织的裂解情况会直接影响到DNA的提取质量,这4种试剂盒在加入裂解液后,水浴30~60 min,此时研磨的粉末都应完全裂解,溶液清澈透明,但在实验过程中,磁珠法试剂盒和DNA快速提取试剂盒在水解1 h后仍然有颗粒状悬浮物。笔者在多次尝试后发现,减少组织用量(毒腺、腹足和肌肉的用量低于20 mg,肝胰脏用量低于10 mg时),裂解情况明显改善;另外,在DNA快速提取试剂盒的裂解液中加入20 μL蛋白酶K,其裂解情况明显改善,最后所提的DNA产率和纯度都有明显的增加;延长裂解时间也有利于组织的裂解,但时间过长(>3 h),会导致DNA产率及纯度的下降。裂解之后的操作原理主要是利用在特定的环境里,核酸能与相应的介质结合,洗涤后将杂质去除,通过更改溶液环境,确保DNA在TE Buffer中溶解[21]。如磁珠法顾名思义是用磁珠吸附DNA,然后用70%乙醇洗涤,此法不涉及有毒性试剂,也不用离心,较适用于自动化操作中;FastPure Tissue DNA提取试剂盒法和海洋动物组织DNA提取试剂盒法均是用滤膜离心柱法,使DNA吸附于硅胶柱上,离心去除杂质,此法主要是离心操作,比较简便;DNA快速提取法是利用DNA在异丙醇中溶解度低的原理,用异丙醇沉淀DNA,操作简单。溶解DNA时用加热至70 ℃TE Buffer,能更好地获取DNA。
迄今为止,已有很多提取芋螺DNA优化方法,如丁青等[22]比较了CTAB法、磁珠法和离心柱法提取菖蒲芋螺(Conus vexillum)3种不同组织器官的DNA的的效果,结果发现,CTAB法在提取大片段DNA时更有效,更适于构建大片段DNA文库,但此法操作比较复杂,且需要使用氯仿等有毒试剂;罗素兰等[23]采用苯酚-蛋白酶k法、缠绕法、快速分离法和改良苯酚-SDS法从6种不同芋螺中提取总DNA,建立了较优化的芋螺DNA提取方法,但这些方法提取腹足组织时很难得到DNA,且不能有效去除蛋白质,因而不适合用来提取腹足DNA。笔者对DNA快速试剂盒进行改良(减少组织用量,裂解时加入蛋白酶K,沉淀DNA时在−20 ℃冰箱中放置20 min),发现用腹足的部位提取的DNA纯度高,产率大,并且因为腹足组织多且易获得,因此,可以用来提取芋螺DNA。
Optimization of Genomic DNA Extraction for Conus litteratus
doi: 10.15886/j.cnki.rdswxb.2020.00.015
- Received Date: 2020-02-26
- Rev Recd Date: 2020-03-05
- Available Online: 2020-07-03
- Publish Date: 2020-06-01
-
Key words:
- Conus litteratus /
- genomic DNA /
- extraction method /
- optimization
Abstract: The acquisition of high-quality genomic DNA is key to cloning of conotoxin genes and gene library construction. Four kits, Magnetic Bead Method Kit, Improved DNA Rapid Extraction Kit, FastPure DNA Isolation Kit, and TIANamp DNA Isolation Kit, were adopted to extract DNA from different tissues and organs of Conus litteratus, and the concentration, purity, size and integrity of the genomic DNA extracted from different tissues and organs by each kit were comprehensively compared. The results showed that the purity and concentration of the genomic DNA extracted with the Improved DNA Rapid Extraction Kit was better. Of the four different organs of C. litterantus, muscular foot had a higher purity in the genomic DNA extracted from its tissues and was the best organ used for extraction of the genomic DNA. The hepatopancreas and venom duct of C. litterantus gave the highest yield of the genomic DNA, but with poor fragment integrity and high contamination. Moreover, specific amplification products with the expected size were obtained through PCR. It is concluded that the best method to acquire the genomic DNA from C. litteratus is to extract the genomic DNA from the muscular foot with the Improved DNA Rapid Extraction Kit, with which high-quality genomic DNA can be produced and meet the requirements of subsequent research.
| Citation: | CHEN Wanyi, LI Xinjia, LUO Sulan, ZHANGSUN Dongting. Optimization of Genomic DNA Extraction for Conus litteratus[J]. Journal of Tropical Biology, 2020, 11(2): 238-244, 250. doi: 10.15886/j.cnki.rdswxb.2020.00.015 |








 DownLoad:
DownLoad: